from pathlib import Path
import itertools as it
import datetime as dt
from dateutil import parser as dtparser
import fsspec
import xarray as xr
import matplotlib.pyplot as plt
import hvplot.xarray
import panel
import echopype as ep
import warnings
warnings.simplefilter("ignore", category=DeprecationWarning)Watching a solar eclipse using an OOI moored echosounder
Jupyter notebook accompanying the manuscript:
Echopype: A Python library for interoperable and scalable processing of ocean sonar data for biological information
Authors: Wu-Jung Lee, Emilio Mayorga, Landung Setiawan, Kavin Nguyen, Imran Majeed, Valentina Staneva
Introduction
Goals
- Illustrate a common workflow for echosounder data conversion, calibration and use. This workflow leverages the standardization applied by echopype. and the power, ease of use and familiarity of libraries in the scientific Python ecosystem.
- Demonstrate the ease to interoperate echosounder data with those from a different instrument in a single computing environment. Without
echopype, additional wrangling across more than one software systems is needed to achieve the same visualization and comparison.
Description
This notebook uses EK60 echosounder data from the U.S. Ocean Observatories Initiative (OOI) to illustrate a common workflow for data conversion, combination, calibration and analysis using echopype, as well as the data interoperability it enables. Without echopype, additional wrangling across more than one software systems is needed to achieve the same visualization and comparison.
We will use data from the OOI Oregon Offshore Cabled Shallow Profiler Mooring collected on August 20-21, 2017. This was the day before and of a solar eclipse, during which the reduced sunlight affected the regular diel vertical migration (DVM) patterns of marine life. This change was directly observed using the upward-looking echosounder mounted on this mooring platform that happened to be within the totality zone. The effect of the solar eclipse was clearly seen by aligning and comparing the echosounder observations with solar radiation data collected by the Bulk Meteorology Instrument Package located on the nearby Coastal Endurance Oregon Offshore Surface Mooring, also maintained by the OOI.
The data used are 19 .raw files with a total volume of approximately 1 GB. With echopype functionality, the raw data files hosted on the OOI Raw Data Archive (an HTTP server) are directly parsed and organized into a standardized representation following in the SONAR-netCDF4 v1.0 convention, and stored to the cloud-optimized Zarr format. The individual converted files are later combined into a single entity that can be easily explored and manipulated.
Outline
- Establish connection with the OOI Raw Data Archive and generate list of target EK60
.rawfiles - Process the archived raw files with
echopype: convert and combine into a single quantity (anEchoDataobject) in a standardized format. - Obtain solar radiation data from an OOI Thredds server.
- Plot the echosounder and solar radiation data together to visualize the zooplankton response to a solar eclipse.
Running the notebook
This notebook can be run with a conda environment created using the conda environment file https://github.com/OSOceanAcoustics/echopype-examples/blob/main/binder/environment.yml. The notebook creates a directory ./exports/notebook3 and save all generated Zarr and netCDF files there.
Warning
The compute_MVBS step in this notebook is not efficient for lazy-loaded data with echopype version 0.6.3. We plan to address this issue soon.
Note
We encourage importing echopype as ep for consistency.
Establish connection with the OOI Raw Data Archive and generate list of target EK60 .raw files
Access and inspect the publicly accessible OOI Raw Data Archive (an HTTP server) as if it were a local file system. This will be done through the Python fsspec file system and bytes storage interface. We will use fsspec.filesystem.glob (fs.glob) to generate a list of all EK60 .raw data files in the archive then filter on file names for target dates of interest.
fs = fsspec.filesystem('https')ooi_raw_url = (
"https://rawdata.oceanobservatories.org/files/"
"CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/2017/08"
)Now let’s specify the range of dates we will be pulling data from. Note that the data filenames contain the time information but were recorded at UTC time.
def in_range(raw_file: str, start: dt.datetime, end: dt.datetime) -> bool:
"""Check if file url is in datetime range"""
file_name = Path(raw_file).name
file_datetime = dtparser.parse(file_name, fuzzy=True)
return file_datetime >= start and file_datetime <= endstart_datetime = dt.datetime(2017, 8, 21, 7, 0)
end_datetime = dt.datetime(2017, 8, 22, 7, 0)On the OOI Raw Data Archive, the monthly folder is further split to daily folders, so we can simply grab data from the desired days.
desired_day_urls = [f"{ooi_raw_url}/{day}" for day in range(start_datetime.day, end_datetime.day + 1)]desired_day_urls['https://rawdata.oceanobservatories.org/files/CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/2017/08/21',
'https://rawdata.oceanobservatories.org/files/CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/2017/08/22']Grab all raw files within daily folders by using the filesytem glob, just like the Linux glob.
all_raw_file_urls = it.chain.from_iterable([fs.glob(f"{day_url}/*.raw") for day_url in desired_day_urls])desired_raw_file_urls = list(filter(
lambda raw_file: in_range(
raw_file,
start_datetime-dt.timedelta(hours=3), # 3 hour buffer to select files
end_datetime+dt.timedelta(hours=3)
),
all_raw_file_urls
))
print(f"There are {len(desired_raw_file_urls)} raw files within the specified datetime range.")There are 19 raw files within the specified datetime range.Process the archived raw files with echopype
Examine the workflow by processing just one file
Let’s first test the echopype workflow by converting and processing 1 file from the above list.
We will use ep.open_raw to directly read in a raw data file from the OOI HTTP server.
The type of sonar needs to be specified as an input argument. The echosounders on the OOI Regional Cabled Array are Simrad EK60 echosounder. All other uncabled echosounders are the Acoustic Zooplankton and Fisher Profiler (AZFP) manufacturered by ASL Environmental Sciences. Echopype supports both of these and other instruments (see echopype documentation for detail).
Converting from raw data files to a standardized data format
Below we already know the path to the 1 file on the http server:
echodata = ep.open_raw(raw_file=desired_raw_file_urls[0], sonar_model="ek60")Here echopype read, parse, and convert content of the raw file into memory, and gives you a nice representation of the converted file below as a Python EchoData object.
echodata-
<xarray.Dataset> Size: 0B Dimensions: () Data variables: *empty* Attributes: conventions: CF-1.7, SONAR-netCDF4-1.0, ACDD-1.3 keywords: EK60 sonar_convention_authority: ICES sonar_convention_name: SONAR-netCDF4 sonar_convention_version: 1.0 summary: title: date_created: 2017-08-21T04:57:17Z -
<xarray.Dataset> Size: 332kB Dimensions: (channel: 3, time1: 5923) Coordinates: * channel (channel) <U39 468B 'GPT 38 kHz 00907208dd13 5-1... * time1 (time1) datetime64[ns] 47kB 2017-08-21T04:57:17.3... Data variables: absorption_indicative (channel, time1) float64 142kB 0.009785 ... 0.05269 sound_speed_indicative (channel, time1) float64 142kB 1.494e+03 ... 1.49... frequency_nominal (channel) float64 24B 3.8e+04 1.2e+05 2e+05 -
<xarray.Dataset> Size: 190kB Dimensions: (time1: 1, time2: 5923, channel: 3) Coordinates: * time1 (time1) datetime64[ns] 8B 2017-08-21T04:57:17.329287 * time2 (time2) datetime64[ns] 47kB 2017-08-21T04:57:17.3292... * channel (channel) <U39 468B 'GPT 38 kHz 00907208dd13 5-1 OO... Data variables: (12/20) latitude (time1) float64 8B nan longitude (time1) float64 8B nan sentence_type (time1) float64 8B nan pitch (time2) float64 47kB 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 roll (time2) float64 47kB 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 vertical_offset (time2) float64 47kB 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 ... ... position_offset_y float64 8B nan position_offset_z float64 8B nan transducer_offset_x (channel) float64 24B 0.0 0.0 0.0 transducer_offset_y (channel) float64 24B 0.0 0.0 0.0 transducer_offset_z (channel) float64 24B 0.0 0.0 0.0 frequency_nominal (channel) float64 24B 3.8e+04 1.2e+05 2e+05 Attributes: platform_name: platform_type: platform_code_ICES: -
<xarray.Dataset> Size: 96B Dimensions: (time1: 1) Coordinates: * time1 (time1) datetime64[ns] 8B 2017-08-21T04:57:17.329287 Data variables: NMEA_datagram (time1) <U22 88B '$SDVLW,0.000,N,0.000,N' Attributes: description: All NMEA sensor datagrams -
<xarray.Dataset> Size: 484B Dimensions: (filenames: 1) Coordinates: * filenames (filenames) int64 8B 0 Data variables: source_filenames (filenames) <U119 476B 'https://rawdata.oceanobservator... Attributes: conversion_software_name: echopype conversion_software_version: 0.8.4.dev58+gae911de conversion_time: 2024-04-26T18:38:27Z -
<xarray.Dataset> Size: 568B Dimensions: (beam_group: 1) Coordinates: * beam_group (beam_group) <U11 44B 'Beam_group1' Data variables: beam_group_descr (beam_group) <U131 524B 'contains backscatter power (un... Attributes: sonar_manufacturer: Simrad sonar_model: EK60 sonar_serial_number: sonar_software_name: ER60 sonar_software_version: 2.4.3 sonar_type: echosounder -
<xarray.Dataset> Size: 229MB Dimensions: (channel: 3, ping_time: 5923, range_sample: 1072) Coordinates: * channel (channel) <U39 468B 'GPT 38 kHz 00907208d... * ping_time (ping_time) datetime64[ns] 47kB 2017-08-21... * range_sample (range_sample) int64 9kB 0 1 2 ... 1070 1071 Data variables: (12/29) frequency_nominal (channel) float64 24B 3.8e+04 1.2e+05 2e+05 beam_type (channel) int64 24B 0 1 0 beamwidth_twoway_alongship (channel) float64 24B 7.1 7.0 7.0 beamwidth_twoway_athwartship (channel) float64 24B 7.1 7.0 7.0 beam_direction_x (channel) float64 24B nan nan nan beam_direction_y (channel) float64 24B nan nan nan ... ... data_type (channel, ping_time) int8 18kB 1 1 1 ... 1 1 sample_time_offset (channel, ping_time) float64 142kB 0.0 ...... channel_mode (channel, ping_time) int8 18kB 0 0 0 ... 0 0 backscatter_r (channel, ping_time, range_sample) float32 76MB ... angle_athwartship (channel, ping_time, range_sample) float32 76MB ... angle_alongship (channel, ping_time, range_sample) float32 76MB ... Attributes: beam_mode: vertical conversion_equation_t: type_3 -
<xarray.Dataset> Size: 892B Dimensions: (channel: 3, pulse_length_bin: 5) Coordinates: * channel (channel) <U39 468B 'GPT 38 kHz 00907208dd13 5-1 OOI.... * pulse_length_bin (pulse_length_bin) int64 40B 0 1 2 3 4 Data variables: frequency_nominal (channel) float64 24B 3.8e+04 1.2e+05 2e+05 sa_correction (channel, pulse_length_bin) float64 120B 0.0 0.0 ... 0.0 gain_correction (channel, pulse_length_bin) float64 120B 24.0 ... 25.0 pulse_length (channel, pulse_length_bin) float64 120B 0.000256 ... ...
The EchoData object can be saved to either the netCDF4 or zarr formats through to_netcdf or to_zarr methods.
# Create directory for files generated in this notebook
base_dpath = Path('./exports/notebook3')
base_dpath.mkdir(exist_ok=True, parents=True)# Create directory for single-raw-file example
output_dpath = Path(base_dpath / 'onefiletest')
output_dpath.mkdir(exist_ok=True)# Save to netCDF format
echodata.to_netcdf(save_path=output_dpath, overwrite=True)# Save to zarr format
echodata.to_zarr(save_path=output_dpath, overwrite=True)Basic echo processing
At present echopype supports basic processing funcionalities including calibration (from raw instrument data records to volume backscattering strength, \(S_V\)), denoising, and computing mean volume backscattering strength, \(\overline{S_V}\) or \(\text{MVBS}\). The Echodata object can be passed into various calibrate and preprocessing functions without having to write out any intermediate files.
Here we demonstrate calibration to obtain \(S_V\). For EK60 data, by default the function uses environmental (sound speed and absorption) and calibration parameters stored in the data file. Users can optionally specify other parameter choices.
# Compute volume backscattering strength (Sv) from raw data
ds_Sv = ep.calibrate.compute_Sv(echodata)The computed Sv is stored with other variables used in the calibration operation.
ds_Sv<xarray.Dataset> Size: 305MB
Dimensions: (channel: 3, ping_time: 5923,
range_sample: 1072, filenames: 1)
Coordinates:
* channel (channel) <U39 468B 'GPT 38 kHz 00907208d...
* ping_time (ping_time) datetime64[ns] 47kB 2017-08-21...
* range_sample (range_sample) int64 9kB 0 1 2 ... 1070 1071
* filenames (filenames) int64 8B 0
Data variables: (12/16)
Sv (channel, ping_time, range_sample) float64 152MB ...
echo_range (channel, ping_time, range_sample) float64 152MB ...
frequency_nominal (channel) float64 24B 3.8e+04 1.2e+05 2e+05
sound_speed (channel, ping_time) float64 142kB 1.494e+...
sound_absorption (channel, ping_time) float64 142kB 0.00978...
sa_correction (channel, ping_time) float64 142kB 0.0 ......
... ...
angle_sensitivity_alongship (channel) float64 24B 21.9 23.0 23.0
angle_sensitivity_athwartship (channel) float64 24B 21.9 23.0 23.0
beamwidth_alongship (channel) float64 24B 7.1 7.0 7.0
beamwidth_athwartship (channel) float64 24B 7.1 7.0 7.0
source_filenames (filenames) <U119 476B 'https://rawdata.oc...
water_level float64 8B 0.0
Attributes:
processing_software_name: echopype
processing_software_version: 0.8.4.dev58+gae911de
processing_time: 2024-04-26T18:38:31Z
processing_function: calibrate.compute_SvQuickly visualize the result
The default xarray visualization functions are useful in getting a quick sense of the data.
First replace the channel dimension and coordinate with the frequency_nominal variable containing actual frequency values. Note that this step is possible only because there are no duplicated frequencies present.
ds_Sv = ep.consolidate.swap_dims_channel_frequency(ds_Sv)ds_Sv.Sv.sel(frequency_nominal=200000).plot.pcolormesh(
x='ping_time', cmap = 'jet', vmin=-80, vmax=-30
);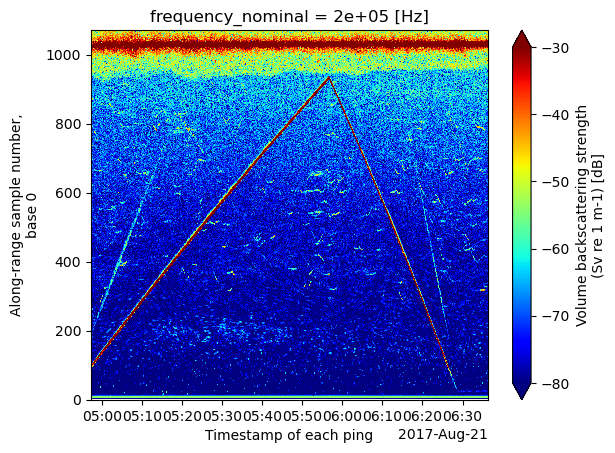
Note that the vertical axis is range_sample. This is the bin (or sample) number as recorded in the data. A separate data variable in ds_Sv contains the physical range (echo_range) from the transducer in meters. echo_range has the same dimension as Sv and may not be uniform across all frequency channels or pings, depending on the echosounder setting during data collection.
Convert multiple files and combine into a single EchoData object
Now that we verified that echopype does work for a single file, let’s proceed to process all sonar data from August 20-21, 2017.
First, convert all desired files from the OOI HTTP server to a local directory.
# Create a directory for all converted files
output_dpath = Path(base_dpath / 'convertallfiles')
output_dpath.mkdir(exist_ok=True)%%time
for raw_file_url in desired_raw_file_urls:
# Read and convert, resulting in echodata object
ed = ep.open_raw(raw_file=raw_file_url, sonar_model='ek60', use_swap=True)
ed.to_zarr(save_path=output_dpath, overwrite=True)Then, assemble a list of EchoData object from the converted files. Note that by using chunks={}, the files are lazy-loaded and only metadata are read into memory, until more operations are executed.
ed_list = []
for converted_file in sorted(output_dpath.glob("*.zarr")):
# ed_list.append(ep.open_converted(converted_file))
ed_list.append(ep.open_converted(converted_file, chunks={}))Combine all the opened files to a single EchoData object.
ed_combined = ep.combine_echodata(ed_list)Below we can inspect this combined EchoData object just as before.
Expand the Provenance group to see how the filenames of all 19 files combined are stored in the coordinate variable echodata_filename, and many more data variables are added to retain information that was previously stored as attributes in the Provenance group of individual EchoData objects.
Expand the Sonar/Beam_group1 to see how data variables such as backscatter_r and angle_alongship contain chunked dask arrays (that are lazy-loadded), instead of regular numpy arrays (that are fully loaded into memory). This means that the computation needed to combine the EchoData objects are specified but not yet executed, which is why the previous line ran through so fast. This is also the mechanism that allows dask to perform its magic for out-of-core computation.
ed_combined-
<xarray.Dataset> Size: 0B Dimensions: () Data variables: *empty* Attributes: conventions: CF-1.7, SONAR-netCDF4-1.0, ACDD-1.3 date_created: 2017-08-21T04:57:17Z keywords: EK60 sonar_convention_authority: ICES sonar_convention_name: SONAR-netCDF4 sonar_convention_version: 1.0 summary: title: -
<xarray.Dataset> Size: 6MB Dimensions: (channel: 3, time1: 109482) Coordinates: * channel (channel) <U39 468B 'GPT 38 kHz 00907208dd13 5-1... * time1 (time1) datetime64[ns] 876kB 2017-08-21T04:57:17.... Data variables: absorption_indicative (channel, time1) float64 3MB dask.array<chunksize=(3, 5923), meta=np.ndarray> frequency_nominal (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> sound_speed_indicative (channel, time1) float64 3MB dask.array<chunksize=(3, 5923), meta=np.ndarray> -
<xarray.Dataset> Size: 4MB Dimensions: (channel: 3, time1: 19, time2: 109478) Coordinates: * time1 (time1) datetime64[ns] 152B 2017-08-21T04:57:17.3292... * channel (channel) <U39 468B 'GPT 38 kHz 00907208dd13 5-1 OO... * time2 (time2) datetime64[ns] 876kB 2017-08-21T04:57:17.329... Data variables: (12/20) MRU_offset_x float64 8B nan MRU_offset_y float64 8B nan MRU_offset_z float64 8B nan MRU_rotation_x float64 8B nan MRU_rotation_y float64 8B nan MRU_rotation_z float64 8B nan ... ... transducer_offset_y (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> transducer_offset_z (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> water_level float64 8B 0.0 pitch (time2) float64 876kB dask.array<chunksize=(5923,), meta=np.ndarray> roll (time2) float64 876kB dask.array<chunksize=(5923,), meta=np.ndarray> vertical_offset (time2) float64 876kB dask.array<chunksize=(5923,), meta=np.ndarray> Attributes: platform_code_ICES: platform_name: platform_type: -
<xarray.Dataset> Size: 8GB Dimensions: (channel: 3, ping_time: 109482, range_sample: 1072) Coordinates: * channel (channel) <U39 468B 'GPT 38 kHz 00907208d... * ping_time (ping_time) datetime64[ns] 876kB 2017-08-2... * range_sample (range_sample) int64 9kB 0 1 2 ... 1070 1071 Data variables: (12/29) angle_alongship (channel, ping_time, range_sample) float64 3GB dask.array<chunksize=(3, 3886, 1072), meta=np.ndarray> angle_athwartship (channel, ping_time, range_sample) float64 3GB dask.array<chunksize=(3, 3886, 1072), meta=np.ndarray> angle_offset_alongship (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> angle_offset_athwartship (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> angle_sensitivity_alongship (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> angle_sensitivity_athwartship (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> ... ... transmit_bandwidth (channel, ping_time) float64 3MB dask.array<chunksize=(3, 5923), meta=np.ndarray> transmit_duration_nominal (channel, ping_time) float64 3MB dask.array<chunksize=(3, 5923), meta=np.ndarray> transmit_frequency_start (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> transmit_frequency_stop (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> transmit_power (channel, ping_time) float64 3MB dask.array<chunksize=(3, 5923), meta=np.ndarray> transmit_type <U2 8B 'CW' Attributes: beam_mode: vertical conversion_equation_t: type_3 -
<xarray.Dataset> Size: 892B Dimensions: (channel: 3, pulse_length_bin: 5) Coordinates: * channel (channel) <U39 468B 'GPT 38 kHz 00907208dd13 5-1 OOI.... * pulse_length_bin (pulse_length_bin) int64 40B 0 1 2 3 4 Data variables: frequency_nominal (channel) float64 24B dask.array<chunksize=(3,), meta=np.ndarray> gain_correction (channel, pulse_length_bin) float64 120B dask.array<chunksize=(3, 5), meta=np.ndarray> pulse_length (channel, pulse_length_bin) float64 120B dask.array<chunksize=(3, 5), meta=np.ndarray> sa_correction (channel, pulse_length_bin) float64 120B dask.array<chunksize=(3, 5), meta=np.ndarray>
Calibrate the combined EchoData and visualize the mean Sv
The single EchoData object is convenient to use for content inspection and calibration. First, compute Sv.
ed_combined.nbytes8475589941.0%%time
ds_Sv = ep.calibrate.compute_Sv(ed_combined).compute()CPU times: user 31.3 s, sys: 44.9 s, total: 1min 16s
Wall time: 36.3 sNext, compute the mean Sv (MVBS) with coherent dimensions along physically meaningful echo_range (in meters) and ping_time from the calibrated data. This processed dataset is easy to visualize. The average bin size along ping_time can be specified using the time series offset alias.
Note that we use .compute() to persist the Sv data in memory in the cell above. This is because the current implementation of compute_MVBS is not efficient for lazy-loaded data. This limitation will be changed in a future release.
%%time
ds_MVBS = ep.commongrid.compute_MVBS(
ds_Sv,
range_bin='0.5m', # 0.5 meters
ping_time_bin='10s' # 10 seconds
)CPU times: user 15.8 s, sys: 15 s, total: 30.8 s
Wall time: 58.4 sThe resulting MVBS Dataset has a coherent echo_range coordinate across all frequencies.
ds_MVBS<xarray.Dataset> Size: 108MB
Dimensions: (channel: 3, ping_time: 11017, echo_range: 410)
Coordinates:
* ping_time (ping_time) datetime64[ns] 88kB 2017-08-21T04:57:10 .....
* channel (channel) <U39 468B 'GPT 38 kHz 00907208dd13 5-1 OOI....
* echo_range (echo_range) float64 3kB 0.0 0.5 1.0 ... 204.0 204.5
Data variables:
Sv (channel, ping_time, echo_range) float64 108MB nan ......
water_level float64 8B 0.0
frequency_nominal (channel) float64 24B 3.8e+04 1.2e+05 2e+05
Attributes:
processing_software_name: echopype
processing_software_version: 0.8.4.dev58+gae911de
processing_time: 2024-04-26T18:40:36Z
processing_function: commongrid.compute_MVBSVisualize MVBS interactively using hvPlot
To visualize, invert the range axis since the echosounder is upward-looking from a platform at approximately 200 m water depth.
ds_MVBS = ds_MVBS.assign_coords(depth=("echo_range", ds_MVBS["echo_range"].values[::-1]))
ds_MVBS = ds_MVBS.swap_dims({'echo_range': 'depth'}) # set depth as data dimensionThen replace the channel dimension and coordinate with the frequency_nominal variable containing actual frequency values. Note that this step is possible only when there are no duplicated frequencies present.
ds_MVBS = ep.consolidate.swap_dims_channel_frequency(ds_MVBS)ds_MVBS["Sv"].sel(frequency_nominal=200000).hvplot.image(
x='ping_time', y='depth',
color='Sv', rasterize=True,
cmap='jet', clim=(-80, -30),
xlabel='Time (UTC)',
ylabel='Depth (m)',
).options(width=800, invert_yaxis=True)/Users/wujung/miniconda3/envs/echopype_examples_20240424/lib/python3.10/site-packages/dask/dataframe/__init__.py:31: FutureWarning:
Dask dataframe query planning is disabled because dask-expr is not installed.
You can install it with `pip install dask[dataframe]` or `conda install dask`.
This will raise in a future version.
warnings.warn(msg, FutureWarning)
OMP: Info #276: omp_set_nested routine deprecated, please use omp_set_max_active_levels instead.Note that the reflection from the sea surface shows up at a location below the depth of 0 m. This is because we have not corrected for the actual depth of the platform on which the echosounder is mounted, and the actual sound speed at the time of data collection (which is related to the calculated range) could also be different from the user-defined sound speed stored in the data file. More accurate platform depth information can be obtained using data from the CTD collocated on the moored platform.
Obtain solar radiation data from an OOI THREDDS server
Now we have the sonar data ready, the next step is to pull solar radiation data collected by a nearby surface mooring also maintained by the OOI. The Bulk Meteorology Instrument Package is located on the Coastal Endurance Oregon Offshore Surface Mooring.
Note: an earlier version of this notebook used the same dataset but pulled from the National Data Buoy Center (NDBC). We thank the Rutgers OOI Data Lab for pointing out the direct data source in one of the data nuggets.
metbk_url = (
"http://thredds.dataexplorer.oceanobservatories.org/thredds/dodsC/ooigoldcopy/public/"
"CE04OSSM-SBD11-06-METBKA000-recovered_host-metbk_a_dcl_instrument_recovered/"
"deployment0004_CE04OSSM-SBD11-06-METBKA000-recovered_host-metbk_a_dcl_instrument_recovered_20170421T022518.003000-20171013T154805.602000.nc#fillmismatch"
)metbk_ds = (
xr.open_dataset(metbk_url)
.swap_dims({'obs': 'time'})
.drop('obs')
.sel(time=slice(start_datetime, end_datetime))[['shortwave_irradiance']]
)
metbk_ds.time.attrs.update({'long_name': 'Time', 'units': 'UTC'})
metbk_ds<xarray.Dataset> Size: 17kB
Dimensions: (time: 1441)
Coordinates:
* time (time) datetime64[ns] 12kB 2017-08-21T07:00:08.2329...
Data variables:
shortwave_irradiance (time) float32 6kB ...
Attributes: (12/73)
node: SBD11
comment:
publisher_email:
sourceUrl: http://oceanobservatories.org/
collection_method: recovered_host
stream: metbk_a_dcl_instrument_recovered
... ...
geospatial_vertical_positive: down
lat: 44.36555
lon: -124.9407
DODS.strlen: 36
DODS.dimName: string36
DODS_EXTRA.Unlimited_Dimension: obsCombine sonar observation with solar radiation measurements
We can finally put everything together and figure out the impact of the eclipse-driven reduction in sunlight on marine zooplankton!
metbk_plot = metbk_ds.hvplot.line(
x='time', y='shortwave_irradiance', responsive=True,
).options(width=800, height=120, logy=True, xlim=(start_datetime, end_datetime))
mvbs_plot = ds_MVBS["Sv"].sel(frequency_nominal=200000, ping_time=slice(start_datetime, end_datetime)).hvplot.image(
x='ping_time', y='depth',
color='Sv', rasterize=True,
cmap='jet', clim=(-80, -30),
xlabel='Time (UTC)',
ylabel='Depth (m)'
).options(width=800, invert_yaxis=True)(metbk_plot + mvbs_plot).cols(1)Look how the dip at solar radiation reading matches exactly with the upwarding moving “blip” at UTC 17:21, August 22, 2017 (local time 10:22 AM). During the solar eclipse, the animals were fooled by the temporary mask of the sun and thought it’s getting dark as at dusk!
Package versions
import datetime
print(f"echopype: {ep.__version__}, xarray: {xr.__version__}, fsspec: {fsspec.__version__}, "
f"hvplot: {hvplot.__version__}")
print(f"\n{datetime.datetime.utcnow()} +00:00")echopype: 0.8.4.dev58+gae911de, xarray: 2024.3.0, fsspec: 2024.3.1, hvplot: 0.9.2
2024-04-26 18:40:44.953619 +00:00