library(sits)SITS - satellite image time series analysis.Loaded sits v1.5.1.
See ?sits for help, citation("sits") for use in publication.
Documentation avaliable in https://e-sensing.github.io/sitsbook/.Update: I was unable to get the code to run when I tried to run this months later.
In this example, I will use sits to prepare some imagery for a specific region and get snow estimates. https://www.usgs.gov/landsat-missions/normalized-difference-snow-index
I will use the Microsoft Planetary Computer data since it has a nice vizualization that will help me find tiles.
First step is to go to the visualizer and figure out the tiles I need. I am looking at an area close to the Canadian border of Washington state. MPC search page. I want to click on the files in the left nav and get the tile ids around “Early Winters”. I need four: “10UFU”, “10UGU”, “10UGV”, “10UFV”.
At first I used tiles, but later went to a small region of interest (roi). When I went from tiles to roi in the sits_regularize() the last date had no data and that caused errors.
I am going to get a month of data for a small region.
roi <- c(
lon_min = -120.436866, lat_min = 47.570978,
lon_max = -120.375207, lat_max = 48.611761
)
s2_cube <- sits_cube(
source = "MPC",
collection = "SENTINEL-2-L2A",
roi = roi,
bands = c("B03", "B11", "CLOUD"),
start_date = as.Date("2022-07-01"),
end_date = as.Date("2022-07-31")
)
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%The time line is not regular.
Warning: cube is not regular, returning all timelines$`10TFT`
[1] "2022-07-01" "2022-07-03" "2022-07-08" "2022-07-11" "2022-07-13"
[6] "2022-07-16" "2022-07-18" "2022-07-21" "2022-07-23" "2022-07-26"
[11] "2022-07-28"
$`10UFU`
[1] "2022-07-01" "2022-07-06" "2022-07-11" "2022-07-16" "2022-07-21"
[6] "2022-07-26"The documentation says things are faster if we save a copy of our files. And we will reduce the area of interest to a really small area and weekly data. This is going to save a bunch of little files (4 tiles x 52 weeks x 3 bands) into the tempdir. This saving takes a long time itself and seems kind of pointless for this case. So I skipped this.
# roi as (lon_min, lon_max, lat_min, lat_max)
roi <- c(
lon_min = -120.436866, lat_min = 48.570978,
lon_max = -120.375207, lat_max = 48.611761
)
sits_cube_copy(s2_cube,
output_dir = here::here("topics-2024/2024-05-24-sits/tempdir/chl1"),
roi = roi
)
|
| | 0%
|
|= | 2%
|
|=== | 4%
|
|==== | 6%
|
|===== | 8%
|
|======= | 10%
|
|======== | 12%
|
|========== | 14%
|
|=========== | 16%
|
|============ | 18%
|
|============== | 20%
|
|=============== | 22%
|
|================ | 24%
|
|================== | 25%
|
|=================== | 27%
|
|===================== | 29%
|
|====================== | 31%
|
|======================= | 33%
|
|========================= | 35%
|
|========================== | 37%
|
|=========================== | 39%
|
|============================= | 41%
|
|============================== | 43%
|
|================================ | 45%
|
|================================= | 47%
|
|================================== | 49%
|
|==================================== | 51%
|
|===================================== | 53%
|
|====================================== | 55%
|
|======================================== | 57%
|
|========================================= | 59%
|
|=========================================== | 61%
|
|============================================ | 63%
|
|============================================= | 65%
|
|=============================================== | 67%
|
|================================================ | 69%
|
|================================================= | 71%
|
|=================================================== | 73%
|
|==================================================== | 75%
|
|====================================================== | 76%
|
|======================================================= | 78%
|
|======================================================== | 80%
|
|========================================================== | 82%
|
|=========================================================== | 84%
|
|============================================================ | 86%
|
|============================================================== | 88%
|
|=============================================================== | 90%
|
|================================================================= | 92%
|
|================================================================== | 94%
|
|=================================================================== | 96%
|
|===================================================================== | 98%
|
|======================================================================| 100%# A tibble: 2 × 12
source collection satellite sensor tile xmin xmax ymin ymax crs
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
1 MPC SENTINEL-2-L2A SENTINEL… MSI 10UFU 688910 693620 5.38e6 5.39e6 EPSG…
2 MPC SENTINEL-2-L2A SENTINEL… MSI 10UFU 688900 693620 5.38e6 5.39e6 EPSG…
# ℹ 2 more variables: labels <list>, file_info <list>Next we need to regularize our cube. I will regularize to 14 day period with a 60m resolution. This is going to take a little while time.
reg_cube <- sits_regularize(
cube = s2_cube,
output_dir = here::here("topics-2024/2024-05-24-sits/tempdir/chl2"),
roi = roi,
res = 60,
period = "P14D",
multicores = 4
)Warning: regularization is faster when data is stored locally
use 'sits_cube_copy()' to copy data locally before regularization
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%https://sentiwiki.copernicus.eu/web/s2-processing#S2Processing-Step1b:NormalisedDifferenceSnowIndex(NDSI)
Make a plot.
-- tmap v3 code detected --[v3->v4] tm_raster(): instead of 'style = "cont"', use 'col.scale = tm_scale_continuous()' and migrate the argument(s) 'style.args', 'midpoint', 'palette' (rename to 'values') to 'tm_scale_continuous(<HERE>)'. For small multiples, specify a 'tm_scale_' for each multiple, and put them in a list: 'col.scale = list(<scale1>, <scale2>, ...)'[v3->v4] tm_raster(): migrate the argument(s) related to the legend of the visual variable 'col', namely 'title' to 'col.legend = tm_legend(<HERE>)'[cols4all] color palettes: use palettes from the R package cols4all. Run 'cols4all::c4a_gui()' to explore them. The old palette name "RdYlGn" is named "rd_yl_gn" (in long format "brewer.rd_yl_gn")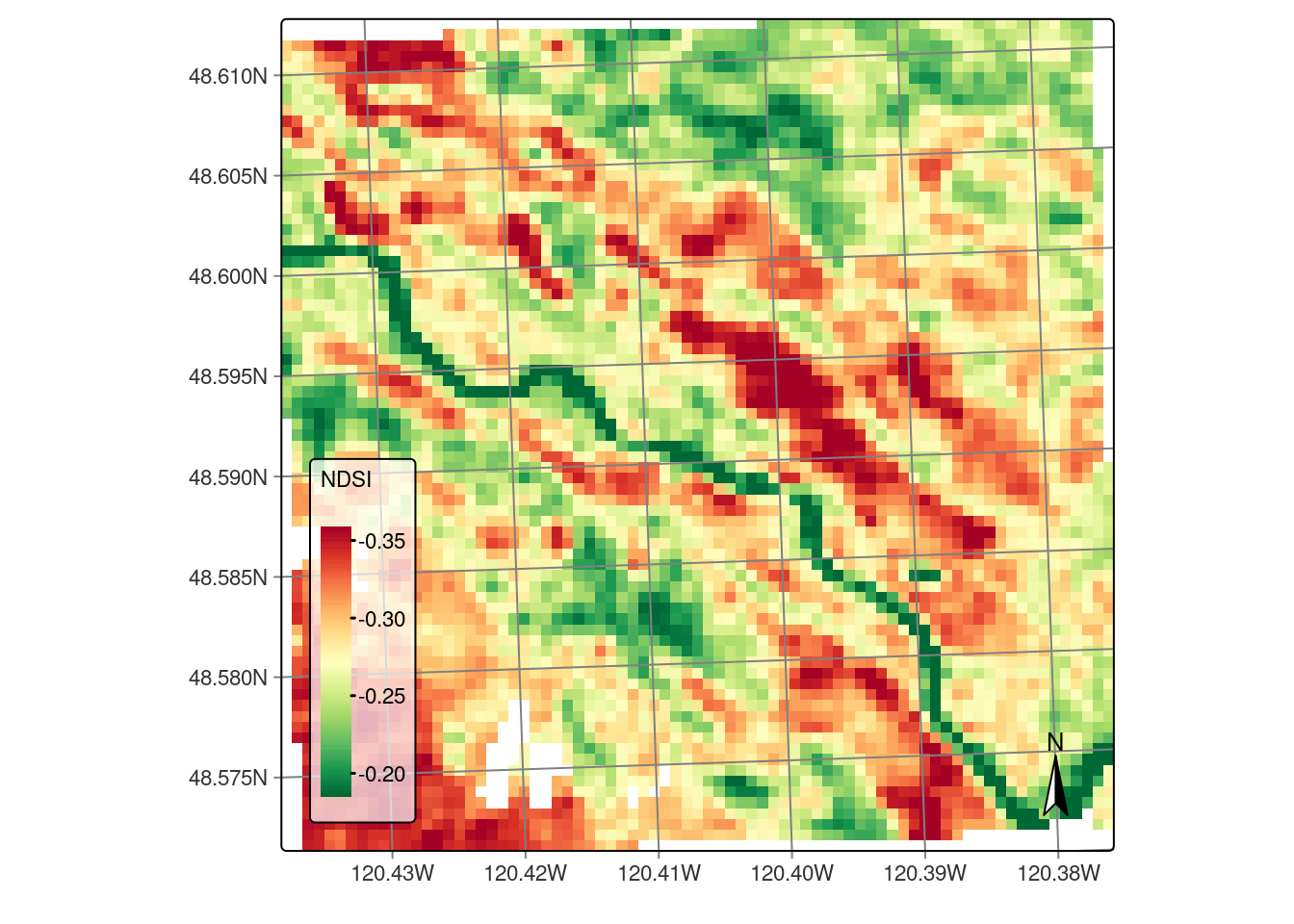
ERROR