📘 Learning Objectives
How to crop a single data file
How to create a data cube (DataSet) with xarray
Extract variables, temporal slices, and spatial slices from an xarray dataset
Summary
In this examples we will use the xarray and earthaccess to subset data and make figures.
For this tutorial we will use the GHRSST Level 4 MUR Global Foundation Sea Surface Temperature Analysis (v4.1) data. This is much higher resolution data than the AVHRR data and we will do spatially subsetting to a small area of interest.
For those not working in the JupyterHub
Create a code cell and run pip install earthaccess
Import Required Packages
# Suppress warnings import warnings'ignore' )'ignore' )from pprint import pprintimport earthaccessimport xarray as xr
Authenticate to NASA Earthdata
We will authenticate our Earthaccess session, and then open the results like we did in the Search & Discovery section.
= earthaccess.login()# are we authenticated? if not auth.authenticated:# ask for credentials and persist them in a .netrc file = "interactive" , persist= True )
Get a vector of urls to our nc files
= 'MUR-JPL-L4-GLOB-v4.1' = "4.1" = "2020-01-01" = "2020-04-01" = (date_start, date_end)# min lon, min lat, max lon, max lat = (- 75.5 , 33.5 , - 73.5 , 35.5 ) = earthaccess.search_data(= short_name,= version,= True ,= date_range,= bbox,len (results)
Crop and plot one netCDF file
Each MUR SST netCDF file is large so I do not want to download. Instead we will subset the data on the server side. We will start with one file.
= earthaccess.open (results[0 :1 ])= xr.open_dataset(fileset[0 ])
<xarray.Dataset> Size: 29GB
Dimensions: (time: 1, lat: 17999, lon: 36000)
Coordinates:
* time (time) datetime64[ns] 8B 2020-01-01T09:00:00
* lat (lat) float32 72kB -89.99 -89.98 -89.97 ... 89.98 89.99
* lon (lon) float32 144kB -180.0 -180.0 -180.0 ... 180.0 180.0
Data variables:
analysed_sst (time, lat, lon) float64 5GB ...
analysis_error (time, lat, lon) float64 5GB ...
mask (time, lat, lon) float32 3GB ...
sea_ice_fraction (time, lat, lon) float64 5GB ...
dt_1km_data (time, lat, lon) timedelta64[ns] 5GB ...
sst_anomaly (time, lat, lon) float64 5GB ...
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid Dimensions: time : 1lat : 17999lon : 36000
Coordinates: (3)
Data variables: (6)
analysed_sst
(time, lat, lon)
float64
...
long_name : analysed sea surface temperature standard_name : sea_surface_foundation_temperature units : kelvin valid_min : -32767 valid_max : 32767 comment : "Final" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF [647964000 values with dtype=float64] analysis_error
(time, lat, lon)
float64
...
long_name : estimated error standard deviation of analysed_sst units : kelvin valid_min : 0 valid_max : 32767 comment : uncertainty in "analysed_sst" [647964000 values with dtype=float64] mask
(time, lat, lon)
float32
...
long_name : sea/land field composite mask valid_min : 1 valid_max : 31 flag_masks : [ 1 2 4 8 16] flag_meanings : open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid comment : mask can be used to further filter the data. source : GMT "grdlandmask", ice flag from sea_ice_fraction data [647964000 values with dtype=float32] sea_ice_fraction
(time, lat, lon)
float64
...
long_name : sea ice area fraction standard_name : sea_ice_area_fraction valid_min : 0 valid_max : 100 source : EUMETSAT OSI-SAF, copyright EUMETSAT comment : ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach. [647964000 values with dtype=float64] dt_1km_data
(time, lat, lon)
timedelta64[ns]
...
long_name : time to most recent 1km data valid_min : -127 valid_max : 127 source : MODIS and VIIRS pixels ingested by MUR comment : The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. "Fill value" indicates absence of such 1km data at the grid point. [647964000 values with dtype=timedelta64[ns]] sst_anomaly
(time, lat, lon)
float64
...
long_name : SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period units : kelvin valid_min : -32767 valid_max : 32767 comment : anomaly reference to the day-of-year average between 2003 and 2014 [647964000 values with dtype=float64] Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2020-01-01 09:00:00'], dtype='datetime64[ns]', name='time', freq=None)) PandasIndex
PandasIndex(Index([-89.98999786376953, -89.9800033569336, -89.97000122070312,
-89.95999908447266, -89.94999694824219, -89.94000244140625,
-89.93000030517578, -89.91999816894531, -89.91000366210938,
-89.9000015258789,
...
89.9000015258789, 89.91000366210938, 89.91999816894531,
89.93000030517578, 89.94000244140625, 89.94999694824219,
89.95999908447266, 89.97000122070312, 89.9800033569336,
89.98999786376953],
dtype='float32', name='lat', length=17999)) PandasIndex
PandasIndex(Index([-179.99000549316406, -179.97999572753906, -179.97000122070312,
-179.9600067138672, -179.9499969482422, -179.94000244140625,
-179.92999267578125, -179.9199981689453, -179.91000366210938,
-179.89999389648438,
...
179.91000366210938, 179.9199981689453, 179.92999267578125,
179.94000244140625, 179.9499969482422, 179.9600067138672,
179.97000122070312, 179.97999572753906, 179.99000549316406,
180.0],
dtype='float32', name='lon', length=36000)) Attributes: (47)
Conventions : CF-1.7 title : Daily MUR SST, Final product summary : A merged, multi-sensor L4 Foundation SST analysis product from JPL. references : http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST institution : Jet Propulsion Laboratory history : created at nominal 4-day latency; replaced nrt (1-day latency) version. comment : MUR = "Multi-scale Ultra-high Resolution" license : These data are available free of charge under data policy of JPL PO.DAAC. id : MUR-JPL-L4-GLOB-v04.1 naming_authority : org.ghrsst product_version : 04.1 uuid : 27665bc0-d5fc-11e1-9b23-0800200c9a66 gds_version_id : 2.0 netcdf_version_id : 4.1 date_created : 20200124T180027Z start_time : 20200101T090000Z stop_time : 20200101T090000Z time_coverage_start : 20191231T210000Z time_coverage_end : 20200101T210000Z file_quality_level : 3 source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF platform : Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships sensor : MODIS, AMSR2, AVHRR, in-situ Metadata_Conventions : Unidata Observation Dataset v1.0 metadata_link : http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1 keywords : Oceans > Ocean Temperature > Sea Surface Temperature keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords standard_name_vocabulary : NetCDF Climate and Forecast (CF) Metadata Convention southernmost_latitude : -90.0 northernmost_latitude : 90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 spatial_resolution : 0.01 degrees geospatial_lat_units : degrees north geospatial_lat_resolution : 0.01 geospatial_lon_units : degrees east geospatial_lon_resolution : 0.01 acknowledgment : Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program. creator_name : JPL MUR SST project creator_email : ghrsst@podaac.jpl.nasa.gov creator_url : http://mur.jpl.nasa.gov project : NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program publisher_name : GHRSST Project Office publisher_url : http://www.ghrsst.org publisher_email : ghrsst-po@nceo.ac.uk processing_level : L4 cdm_data_type : grid
Note that xarray works with “lazy” computation whenever possible. In this case, the metadata are loaded into JupyterHub memory, but the data arrays and their values are not — until there is a need for them.
Let’s print out all the variable names.
for v in ds.variables:print (v)
time
lat
lon
analysed_sst
analysis_error
mask
sea_ice_fraction
dt_1km_data
sst_anomaly
Of the variables listed above, we are interested in analysed_sst.
'analysed_sst' ].attrs
{'long_name': 'analysed sea surface temperature',
'standard_name': 'sea_surface_foundation_temperature',
'units': 'kelvin',
'valid_min': -32767,
'valid_max': 32767,
'comment': '"Final" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation',
'source': 'MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF'}
Subsetting
In addition to directly accessing the files archived and distributed by each of the NASA DAACs, many datasets also support services that allow us to customize the data via subsetting, reformatting, reprojection/regridding, and file aggregation. What does subsetting mean? To subset means to extract only the portions of a dataset that are needed for a given purpose.
There are three primary types of subsetting that we will walk through: 1. Temporal 2. Spatial 3. Variable
In each case, we will be excluding parts of the dataset that are not wanted using xarray. Note that “subsetting” is also called a data “transformation”.
# Display the full dataset's metadata
<xarray.Dataset> Size: 29GB
Dimensions: (time: 1, lat: 17999, lon: 36000)
Coordinates:
* time (time) datetime64[ns] 8B 2020-01-01T09:00:00
* lat (lat) float32 72kB -89.99 -89.98 -89.97 ... 89.98 89.99
* lon (lon) float32 144kB -180.0 -180.0 -180.0 ... 180.0 180.0
Data variables:
analysed_sst (time, lat, lon) float64 5GB ...
analysis_error (time, lat, lon) float64 5GB ...
mask (time, lat, lon) float32 3GB ...
sea_ice_fraction (time, lat, lon) float64 5GB ...
dt_1km_data (time, lat, lon) timedelta64[ns] 5GB ...
sst_anomaly (time, lat, lon) float64 5GB ...
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid Dimensions: time : 1lat : 17999lon : 36000
Coordinates: (3)
Data variables: (6)
analysed_sst
(time, lat, lon)
float64
...
long_name : analysed sea surface temperature standard_name : sea_surface_foundation_temperature units : kelvin valid_min : -32767 valid_max : 32767 comment : "Final" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF [647964000 values with dtype=float64] analysis_error
(time, lat, lon)
float64
...
long_name : estimated error standard deviation of analysed_sst units : kelvin valid_min : 0 valid_max : 32767 comment : uncertainty in "analysed_sst" [647964000 values with dtype=float64] mask
(time, lat, lon)
float32
...
long_name : sea/land field composite mask valid_min : 1 valid_max : 31 flag_masks : [ 1 2 4 8 16] flag_meanings : open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid comment : mask can be used to further filter the data. source : GMT "grdlandmask", ice flag from sea_ice_fraction data [647964000 values with dtype=float32] sea_ice_fraction
(time, lat, lon)
float64
...
long_name : sea ice area fraction standard_name : sea_ice_area_fraction valid_min : 0 valid_max : 100 source : EUMETSAT OSI-SAF, copyright EUMETSAT comment : ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach. [647964000 values with dtype=float64] dt_1km_data
(time, lat, lon)
timedelta64[ns]
...
long_name : time to most recent 1km data valid_min : -127 valid_max : 127 source : MODIS and VIIRS pixels ingested by MUR comment : The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. "Fill value" indicates absence of such 1km data at the grid point. [647964000 values with dtype=timedelta64[ns]] sst_anomaly
(time, lat, lon)
float64
...
long_name : SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period units : kelvin valid_min : -32767 valid_max : 32767 comment : anomaly reference to the day-of-year average between 2003 and 2014 [647964000 values with dtype=float64] Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2020-01-01 09:00:00'], dtype='datetime64[ns]', name='time', freq=None)) PandasIndex
PandasIndex(Index([-89.98999786376953, -89.9800033569336, -89.97000122070312,
-89.95999908447266, -89.94999694824219, -89.94000244140625,
-89.93000030517578, -89.91999816894531, -89.91000366210938,
-89.9000015258789,
...
89.9000015258789, 89.91000366210938, 89.91999816894531,
89.93000030517578, 89.94000244140625, 89.94999694824219,
89.95999908447266, 89.97000122070312, 89.9800033569336,
89.98999786376953],
dtype='float32', name='lat', length=17999)) PandasIndex
PandasIndex(Index([-179.99000549316406, -179.97999572753906, -179.97000122070312,
-179.9600067138672, -179.9499969482422, -179.94000244140625,
-179.92999267578125, -179.9199981689453, -179.91000366210938,
-179.89999389648438,
...
179.91000366210938, 179.9199981689453, 179.92999267578125,
179.94000244140625, 179.9499969482422, 179.9600067138672,
179.97000122070312, 179.97999572753906, 179.99000549316406,
180.0],
dtype='float32', name='lon', length=36000)) Attributes: (47)
Conventions : CF-1.7 title : Daily MUR SST, Final product summary : A merged, multi-sensor L4 Foundation SST analysis product from JPL. references : http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST institution : Jet Propulsion Laboratory history : created at nominal 4-day latency; replaced nrt (1-day latency) version. comment : MUR = "Multi-scale Ultra-high Resolution" license : These data are available free of charge under data policy of JPL PO.DAAC. id : MUR-JPL-L4-GLOB-v04.1 naming_authority : org.ghrsst product_version : 04.1 uuid : 27665bc0-d5fc-11e1-9b23-0800200c9a66 gds_version_id : 2.0 netcdf_version_id : 4.1 date_created : 20200124T180027Z start_time : 20200101T090000Z stop_time : 20200101T090000Z time_coverage_start : 20191231T210000Z time_coverage_end : 20200101T210000Z file_quality_level : 3 source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF platform : Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships sensor : MODIS, AMSR2, AVHRR, in-situ Metadata_Conventions : Unidata Observation Dataset v1.0 metadata_link : http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1 keywords : Oceans > Ocean Temperature > Sea Surface Temperature keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords standard_name_vocabulary : NetCDF Climate and Forecast (CF) Metadata Convention southernmost_latitude : -90.0 northernmost_latitude : 90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 spatial_resolution : 0.01 degrees geospatial_lat_units : degrees north geospatial_lat_resolution : 0.01 geospatial_lon_units : degrees east geospatial_lon_resolution : 0.01 acknowledgment : Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program. creator_name : JPL MUR SST project creator_email : ghrsst@podaac.jpl.nasa.gov creator_url : http://mur.jpl.nasa.gov project : NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program publisher_name : GHRSST Project Office publisher_url : http://www.ghrsst.org publisher_email : ghrsst-po@nceo.ac.uk processing_level : L4 cdm_data_type : grid
Now we will prepare a subset. We’re using essentially the same spatial bounds as above; however, as opposed to the earthaccess inputs above, here we must provide inputs in the formats expected by xarray. Instead of a single, four-element, bounding box, we use Python slice objects, which are defined by starting and ending numbers.
= ds.sel(time= date_start, lat= slice (33.5 , 35.5 ), lon= slice (- 75.5 , - 73.5 ))
<xarray.Dataset> Size: 2MB
Dimensions: (time: 1, lat: 201, lon: 201)
Coordinates:
* time (time) datetime64[ns] 8B 2020-01-01T09:00:00
* lat (lat) float32 804B 33.5 33.51 33.52 ... 35.48 35.49 35.5
* lon (lon) float32 804B -75.5 -75.49 -75.48 ... -73.51 -73.5
Data variables:
analysed_sst (time, lat, lon) float64 323kB ...
analysis_error (time, lat, lon) float64 323kB ...
mask (time, lat, lon) float32 162kB ...
sea_ice_fraction (time, lat, lon) float64 323kB ...
dt_1km_data (time, lat, lon) timedelta64[ns] 323kB ...
sst_anomaly (time, lat, lon) float64 323kB ...
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid Dimensions:
Coordinates: (3)
Data variables: (6)
analysed_sst
(time, lat, lon)
float64
...
long_name : analysed sea surface temperature standard_name : sea_surface_foundation_temperature units : kelvin valid_min : -32767 valid_max : 32767 comment : "Final" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF [40401 values with dtype=float64] analysis_error
(time, lat, lon)
float64
...
long_name : estimated error standard deviation of analysed_sst units : kelvin valid_min : 0 valid_max : 32767 comment : uncertainty in "analysed_sst" [40401 values with dtype=float64] mask
(time, lat, lon)
float32
...
long_name : sea/land field composite mask valid_min : 1 valid_max : 31 flag_masks : [ 1 2 4 8 16] flag_meanings : open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid comment : mask can be used to further filter the data. source : GMT "grdlandmask", ice flag from sea_ice_fraction data [40401 values with dtype=float32] sea_ice_fraction
(time, lat, lon)
float64
...
long_name : sea ice area fraction standard_name : sea_ice_area_fraction valid_min : 0 valid_max : 100 source : EUMETSAT OSI-SAF, copyright EUMETSAT comment : ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach. [40401 values with dtype=float64] dt_1km_data
(time, lat, lon)
timedelta64[ns]
...
long_name : time to most recent 1km data valid_min : -127 valid_max : 127 source : MODIS and VIIRS pixels ingested by MUR comment : The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. "Fill value" indicates absence of such 1km data at the grid point. [40401 values with dtype=timedelta64[ns]] sst_anomaly
(time, lat, lon)
float64
...
long_name : SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period units : kelvin valid_min : -32767 valid_max : 32767 comment : anomaly reference to the day-of-year average between 2003 and 2014 [40401 values with dtype=float64] Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2020-01-01 09:00:00'], dtype='datetime64[ns]', name='time', freq=None)) PandasIndex
PandasIndex(Index([ 33.5, 33.5099983215332, 33.52000045776367,
33.529998779296875, 33.540000915527344, 33.54999923706055,
33.560001373291016, 33.56999969482422, 33.58000183105469,
33.59000015258789,
...
35.40999984741211, 35.41999816894531, 35.43000030517578,
35.439998626708984, 35.45000076293945, 35.459999084472656,
35.470001220703125, 35.47999954223633, 35.4900016784668,
35.5],
dtype='float32', name='lat', length=201)) PandasIndex
PandasIndex(Index([ -75.5, -75.48999786376953, -75.4800033569336,
-75.47000122070312, -75.45999908447266, -75.44999694824219,
-75.44000244140625, -75.43000030517578, -75.41999816894531,
-75.41000366210938,
...
-73.58999633789062, -73.58000183105469, -73.56999969482422,
-73.55999755859375, -73.55000305175781, -73.54000091552734,
-73.52999877929688, -73.5199966430664, -73.51000213623047,
-73.5],
dtype='float32', name='lon', length=201)) Attributes: (47)
Conventions : CF-1.7 title : Daily MUR SST, Final product summary : A merged, multi-sensor L4 Foundation SST analysis product from JPL. references : http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST institution : Jet Propulsion Laboratory history : created at nominal 4-day latency; replaced nrt (1-day latency) version. comment : MUR = "Multi-scale Ultra-high Resolution" license : These data are available free of charge under data policy of JPL PO.DAAC. id : MUR-JPL-L4-GLOB-v04.1 naming_authority : org.ghrsst product_version : 04.1 uuid : 27665bc0-d5fc-11e1-9b23-0800200c9a66 gds_version_id : 2.0 netcdf_version_id : 4.1 date_created : 20200124T180027Z start_time : 20200101T090000Z stop_time : 20200101T090000Z time_coverage_start : 20191231T210000Z time_coverage_end : 20200101T210000Z file_quality_level : 3 source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF platform : Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships sensor : MODIS, AMSR2, AVHRR, in-situ Metadata_Conventions : Unidata Observation Dataset v1.0 metadata_link : http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1 keywords : Oceans > Ocean Temperature > Sea Surface Temperature keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords standard_name_vocabulary : NetCDF Climate and Forecast (CF) Metadata Convention southernmost_latitude : -90.0 northernmost_latitude : 90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 spatial_resolution : 0.01 degrees geospatial_lat_units : degrees north geospatial_lat_resolution : 0.01 geospatial_lon_units : degrees east geospatial_lon_resolution : 0.01 acknowledgment : Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program. creator_name : JPL MUR SST project creator_email : ghrsst@podaac.jpl.nasa.gov creator_url : http://mur.jpl.nasa.gov project : NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program publisher_name : GHRSST Project Office publisher_url : http://www.ghrsst.org publisher_email : ghrsst-po@nceo.ac.uk processing_level : L4 cdm_data_type : grid
Plotting
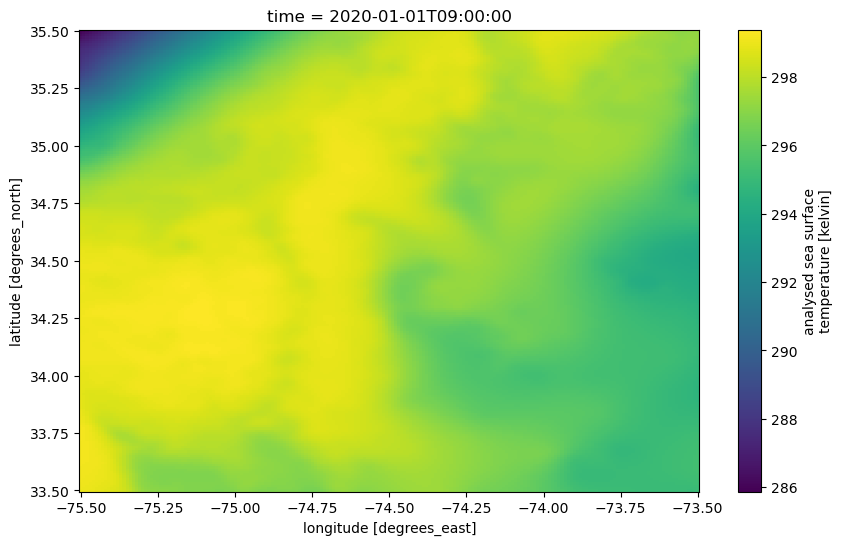
We will first plot using the methods built-in to the xarray package.
Note that, as opposed to the “lazy” loading of metadata previously, this will now perform “eager” computation, pulling the required data chunks.
'analysed_sst' ].plot(figsize= (10 ,6 ), x= 'lon' , y= 'lat' );
Create a data cube by combining multiple netCDF files
When we open multiple files, we use open_mfdataset(). Once again, we are doing lazy loading. Note this method works best if you are in the same Amazon Web Services (AWS) region as the data (us-west-2) and can use S3 connection. For the EDM workshop, we are on an Azure JupyterHub and are using https connection so this is much much slower. If we had spun up this JupyterHub on AWS us-west-2 where the NASA data are hosted, we could load a whole year of data instantly. We will load just a few days so it doesn’t take so long.
= earthaccess.open (results[0 :6 ])= xr.open_mfdataset(fileset[0 :5 ])
<xarray.Dataset> Size: 143GB
Dimensions: (time: 5, lat: 17999, lon: 36000)
Coordinates:
* time (time) datetime64[ns] 40B 2020-01-01T09:00:00 ... 2020-...
* lat (lat) float32 72kB -89.99 -89.98 -89.97 ... 89.98 89.99
* lon (lon) float32 144kB -180.0 -180.0 -180.0 ... 180.0 180.0
Data variables:
analysed_sst (time, lat, lon) float64 26GB dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
analysis_error (time, lat, lon) float64 26GB dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
mask (time, lat, lon) float32 13GB dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
sea_ice_fraction (time, lat, lon) float64 26GB dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
dt_1km_data (time, lat, lon) timedelta64[ns] 26GB dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
sst_anomaly (time, lat, lon) float64 26GB dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid Dimensions: time : 5lat : 17999lon : 36000
Coordinates: (3)
time
(time)
datetime64[ns]
2020-01-01T09:00:00 ... 2020-01-...
long_name : reference time of sst field standard_name : time axis : T comment : Nominal time of analyzed fields array(['2020-01-01T09:00:00.000000000', '2020-01-02T09:00:00.000000000',
'2020-01-03T09:00:00.000000000', '2020-01-04T09:00:00.000000000',
'2020-01-05T09:00:00.000000000'], dtype='datetime64[ns]') lat
(lat)
float32
-89.99 -89.98 ... 89.98 89.99
long_name : latitude standard_name : latitude axis : Y units : degrees_north valid_min : -90.0 valid_max : 90.0 comment : geolocations inherited from the input data without correction array([-89.99, -89.98, -89.97, ..., 89.97, 89.98, 89.99], dtype=float32) lon
(lon)
float32
-180.0 -180.0 ... 180.0 180.0
long_name : longitude standard_name : longitude axis : X units : degrees_east valid_min : -180.0 valid_max : 180.0 comment : geolocations inherited from the input data without correction array([-179.99, -179.98, -179.97, ..., 179.98, 179.99, 180. ],
dtype=float32) Data variables: (6)
analysed_sst
(time, lat, lon)
float64
dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
long_name : analysed sea surface temperature standard_name : sea_surface_foundation_temperature units : kelvin valid_min : -32767 valid_max : 32767 comment : "Final" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
Bytes
24.14 GiB
15.98 MiB
Shape
(5, 17999, 36000)
(1, 1023, 2047)
Dask graph
1620 chunks in 11 graph layers
Data type
float64 numpy.ndarray
36000 17999 5
analysis_error
(time, lat, lon)
float64
dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
long_name : estimated error standard deviation of analysed_sst units : kelvin valid_min : 0 valid_max : 32767 comment : uncertainty in "analysed_sst"
Bytes
24.14 GiB
15.98 MiB
Shape
(5, 17999, 36000)
(1, 1023, 2047)
Dask graph
1620 chunks in 11 graph layers
Data type
float64 numpy.ndarray
36000 17999 5
mask
(time, lat, lon)
float32
dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
long_name : sea/land field composite mask valid_min : 1 valid_max : 31 flag_masks : [ 1 2 4 8 16] flag_meanings : open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid comment : mask can be used to further filter the data. source : GMT "grdlandmask", ice flag from sea_ice_fraction data
Bytes
12.07 GiB
15.98 MiB
Shape
(5, 17999, 36000)
(1, 1447, 2895)
Dask graph
845 chunks in 11 graph layers
Data type
float32 numpy.ndarray
36000 17999 5
sea_ice_fraction
(time, lat, lon)
float64
dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
long_name : sea ice area fraction standard_name : sea_ice_area_fraction valid_min : 0 valid_max : 100 source : EUMETSAT OSI-SAF, copyright EUMETSAT comment : ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach.
Bytes
24.14 GiB
31.96 MiB
Shape
(5, 17999, 36000)
(1, 1447, 2895)
Dask graph
845 chunks in 11 graph layers
Data type
float64 numpy.ndarray
36000 17999 5
dt_1km_data
(time, lat, lon)
timedelta64[ns]
dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
long_name : time to most recent 1km data valid_min : -127 valid_max : 127 source : MODIS and VIIRS pixels ingested by MUR comment : The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. "Fill value" indicates absence of such 1km data at the grid point.
Bytes
24.14 GiB
31.96 MiB
Shape
(5, 17999, 36000)
(1, 1447, 2895)
Dask graph
845 chunks in 11 graph layers
Data type
timedelta64[ns] numpy.ndarray
36000 17999 5
sst_anomaly
(time, lat, lon)
float64
dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
long_name : SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period units : kelvin valid_min : -32767 valid_max : 32767 comment : anomaly reference to the day-of-year average between 2003 and 2014
Bytes
24.14 GiB
15.98 MiB
Shape
(5, 17999, 36000)
(1, 1023, 2047)
Dask graph
1620 chunks in 11 graph layers
Data type
float64 numpy.ndarray
36000 17999 5
Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2020-01-01 09:00:00', '2020-01-02 09:00:00',
'2020-01-03 09:00:00', '2020-01-04 09:00:00',
'2020-01-05 09:00:00'],
dtype='datetime64[ns]', name='time', freq=None)) PandasIndex
PandasIndex(Index([-89.98999786376953, -89.9800033569336, -89.97000122070312,
-89.95999908447266, -89.94999694824219, -89.94000244140625,
-89.93000030517578, -89.91999816894531, -89.91000366210938,
-89.9000015258789,
...
89.9000015258789, 89.91000366210938, 89.91999816894531,
89.93000030517578, 89.94000244140625, 89.94999694824219,
89.95999908447266, 89.97000122070312, 89.9800033569336,
89.98999786376953],
dtype='float32', name='lat', length=17999)) PandasIndex
PandasIndex(Index([-179.99000549316406, -179.97999572753906, -179.97000122070312,
-179.9600067138672, -179.9499969482422, -179.94000244140625,
-179.92999267578125, -179.9199981689453, -179.91000366210938,
-179.89999389648438,
...
179.91000366210938, 179.9199981689453, 179.92999267578125,
179.94000244140625, 179.9499969482422, 179.9600067138672,
179.97000122070312, 179.97999572753906, 179.99000549316406,
180.0],
dtype='float32', name='lon', length=36000)) Attributes: (47)
Conventions : CF-1.7 title : Daily MUR SST, Final product summary : A merged, multi-sensor L4 Foundation SST analysis product from JPL. references : http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST institution : Jet Propulsion Laboratory history : created at nominal 4-day latency; replaced nrt (1-day latency) version. comment : MUR = "Multi-scale Ultra-high Resolution" license : These data are available free of charge under data policy of JPL PO.DAAC. id : MUR-JPL-L4-GLOB-v04.1 naming_authority : org.ghrsst product_version : 04.1 uuid : 27665bc0-d5fc-11e1-9b23-0800200c9a66 gds_version_id : 2.0 netcdf_version_id : 4.1 date_created : 20200124T180027Z start_time : 20200101T090000Z stop_time : 20200101T090000Z time_coverage_start : 20191231T210000Z time_coverage_end : 20200101T210000Z file_quality_level : 3 source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF platform : Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships sensor : MODIS, AMSR2, AVHRR, in-situ Metadata_Conventions : Unidata Observation Dataset v1.0 metadata_link : http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1 keywords : Oceans > Ocean Temperature > Sea Surface Temperature keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords standard_name_vocabulary : NetCDF Climate and Forecast (CF) Metadata Convention southernmost_latitude : -90.0 northernmost_latitude : 90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 spatial_resolution : 0.01 degrees geospatial_lat_units : degrees north geospatial_lat_resolution : 0.01 geospatial_lon_units : degrees east geospatial_lon_resolution : 0.01 acknowledgment : Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program. creator_name : JPL MUR SST project creator_email : ghrsst@podaac.jpl.nasa.gov creator_url : http://mur.jpl.nasa.gov project : NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program publisher_name : GHRSST Project Office publisher_url : http://www.ghrsst.org publisher_email : ghrsst-po@nceo.ac.uk processing_level : L4 cdm_data_type : grid
We can subset a spatial box as we did with a single file.
= ds.sel(lat= slice (33.5 , 35.5 ), lon= slice (- 75.5 , - 73.5 ))
<xarray.Dataset> Size: 9MB
Dimensions: (time: 5, lat: 201, lon: 201)
Coordinates:
* time (time) datetime64[ns] 40B 2020-01-01T09:00:00 ... 2020-...
* lat (lat) float32 804B 33.5 33.51 33.52 ... 35.48 35.49 35.5
* lon (lon) float32 804B -75.5 -75.49 -75.48 ... -73.51 -73.5
Data variables:
analysed_sst (time, lat, lon) float64 2MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
analysis_error (time, lat, lon) float64 2MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
mask (time, lat, lon) float32 808kB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
sea_ice_fraction (time, lat, lon) float64 2MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
dt_1km_data (time, lat, lon) timedelta64[ns] 2MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
sst_anomaly (time, lat, lon) float64 2MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid Dimensions:
Coordinates: (3)
time
(time)
datetime64[ns]
2020-01-01T09:00:00 ... 2020-01-...
long_name : reference time of sst field standard_name : time axis : T comment : Nominal time of analyzed fields array(['2020-01-01T09:00:00.000000000', '2020-01-02T09:00:00.000000000',
'2020-01-03T09:00:00.000000000', '2020-01-04T09:00:00.000000000',
'2020-01-05T09:00:00.000000000'], dtype='datetime64[ns]') lat
(lat)
float32
33.5 33.51 33.52 ... 35.49 35.5
long_name : latitude standard_name : latitude axis : Y units : degrees_north valid_min : -90.0 valid_max : 90.0 comment : geolocations inherited from the input data without correction array([33.5 , 33.51, 33.52, ..., 35.48, 35.49, 35.5 ], dtype=float32) lon
(lon)
float32
-75.5 -75.49 ... -73.51 -73.5
long_name : longitude standard_name : longitude axis : X units : degrees_east valid_min : -180.0 valid_max : 180.0 comment : geolocations inherited from the input data without correction array([-75.5 , -75.49, -75.48, ..., -73.52, -73.51, -73.5 ], dtype=float32) Data variables: (6)
analysed_sst
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : analysed sea surface temperature standard_name : sea_surface_foundation_temperature units : kelvin valid_min : -32767 valid_max : 32767 comment : "Final" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
Bytes
1.54 MiB
315.63 kiB
Shape
(5, 201, 201)
(1, 201, 201)
Dask graph
5 chunks in 12 graph layers
Data type
float64 numpy.ndarray
201 201 5
analysis_error
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : estimated error standard deviation of analysed_sst units : kelvin valid_min : 0 valid_max : 32767 comment : uncertainty in "analysed_sst"
Bytes
1.54 MiB
315.63 kiB
Shape
(5, 201, 201)
(1, 201, 201)
Dask graph
5 chunks in 12 graph layers
Data type
float64 numpy.ndarray
201 201 5
mask
(time, lat, lon)
float32
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : sea/land field composite mask valid_min : 1 valid_max : 31 flag_masks : [ 1 2 4 8 16] flag_meanings : open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid comment : mask can be used to further filter the data. source : GMT "grdlandmask", ice flag from sea_ice_fraction data
Bytes
789.08 kiB
157.82 kiB
Shape
(5, 201, 201)
(1, 201, 201)
Dask graph
5 chunks in 12 graph layers
Data type
float32 numpy.ndarray
201 201 5
sea_ice_fraction
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : sea ice area fraction standard_name : sea_ice_area_fraction valid_min : 0 valid_max : 100 source : EUMETSAT OSI-SAF, copyright EUMETSAT comment : ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach.
Bytes
1.54 MiB
315.63 kiB
Shape
(5, 201, 201)
(1, 201, 201)
Dask graph
5 chunks in 12 graph layers
Data type
float64 numpy.ndarray
201 201 5
dt_1km_data
(time, lat, lon)
timedelta64[ns]
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : time to most recent 1km data valid_min : -127 valid_max : 127 source : MODIS and VIIRS pixels ingested by MUR comment : The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. "Fill value" indicates absence of such 1km data at the grid point.
Bytes
1.54 MiB
315.63 kiB
Shape
(5, 201, 201)
(1, 201, 201)
Dask graph
5 chunks in 12 graph layers
Data type
timedelta64[ns] numpy.ndarray
201 201 5
sst_anomaly
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period units : kelvin valid_min : -32767 valid_max : 32767 comment : anomaly reference to the day-of-year average between 2003 and 2014
Bytes
1.54 MiB
315.63 kiB
Shape
(5, 201, 201)
(1, 201, 201)
Dask graph
5 chunks in 12 graph layers
Data type
float64 numpy.ndarray
201 201 5
Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2020-01-01 09:00:00', '2020-01-02 09:00:00',
'2020-01-03 09:00:00', '2020-01-04 09:00:00',
'2020-01-05 09:00:00'],
dtype='datetime64[ns]', name='time', freq=None)) PandasIndex
PandasIndex(Index([ 33.5, 33.5099983215332, 33.52000045776367,
33.529998779296875, 33.540000915527344, 33.54999923706055,
33.560001373291016, 33.56999969482422, 33.58000183105469,
33.59000015258789,
...
35.40999984741211, 35.41999816894531, 35.43000030517578,
35.439998626708984, 35.45000076293945, 35.459999084472656,
35.470001220703125, 35.47999954223633, 35.4900016784668,
35.5],
dtype='float32', name='lat', length=201)) PandasIndex
PandasIndex(Index([ -75.5, -75.48999786376953, -75.4800033569336,
-75.47000122070312, -75.45999908447266, -75.44999694824219,
-75.44000244140625, -75.43000030517578, -75.41999816894531,
-75.41000366210938,
...
-73.58999633789062, -73.58000183105469, -73.56999969482422,
-73.55999755859375, -73.55000305175781, -73.54000091552734,
-73.52999877929688, -73.5199966430664, -73.51000213623047,
-73.5],
dtype='float32', name='lon', length=201)) Attributes: (47)
Conventions : CF-1.7 title : Daily MUR SST, Final product summary : A merged, multi-sensor L4 Foundation SST analysis product from JPL. references : http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST institution : Jet Propulsion Laboratory history : created at nominal 4-day latency; replaced nrt (1-day latency) version. comment : MUR = "Multi-scale Ultra-high Resolution" license : These data are available free of charge under data policy of JPL PO.DAAC. id : MUR-JPL-L4-GLOB-v04.1 naming_authority : org.ghrsst product_version : 04.1 uuid : 27665bc0-d5fc-11e1-9b23-0800200c9a66 gds_version_id : 2.0 netcdf_version_id : 4.1 date_created : 20200124T180027Z start_time : 20200101T090000Z stop_time : 20200101T090000Z time_coverage_start : 20191231T210000Z time_coverage_end : 20200101T210000Z file_quality_level : 3 source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF platform : Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships sensor : MODIS, AMSR2, AVHRR, in-situ Metadata_Conventions : Unidata Observation Dataset v1.0 metadata_link : http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1 keywords : Oceans > Ocean Temperature > Sea Surface Temperature keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords standard_name_vocabulary : NetCDF Climate and Forecast (CF) Metadata Convention southernmost_latitude : -90.0 northernmost_latitude : 90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 spatial_resolution : 0.01 degrees geospatial_lat_units : degrees north geospatial_lat_resolution : 0.01 geospatial_lon_units : degrees east geospatial_lon_resolution : 0.01 acknowledgment : Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program. creator_name : JPL MUR SST project creator_email : ghrsst@podaac.jpl.nasa.gov creator_url : http://mur.jpl.nasa.gov project : NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program publisher_name : GHRSST Project Office publisher_url : http://www.ghrsst.org publisher_email : ghrsst-po@nceo.ac.uk processing_level : L4 cdm_data_type : grid
We can subset a slice of days also.
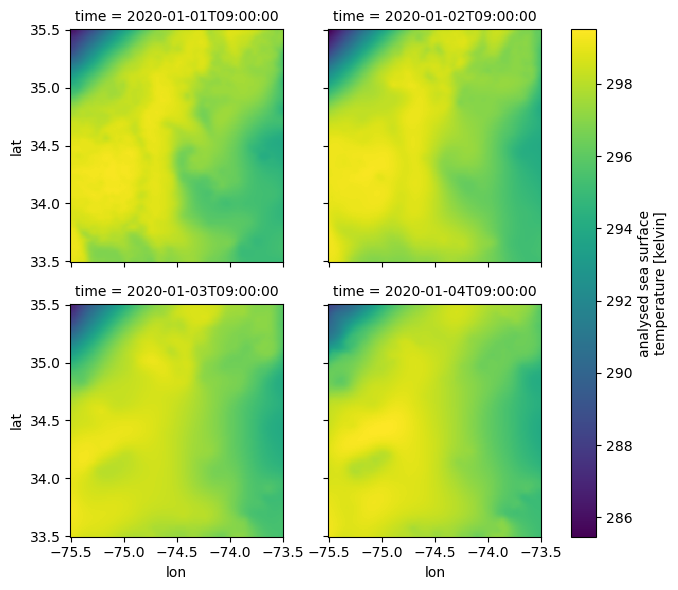
= ds_subset.sel(time= slice ("2020-01-01" , "2020-01-04" ))
<xarray.Dataset> Size: 7MB
Dimensions: (time: 4, lat: 201, lon: 201)
Coordinates:
* time (time) datetime64[ns] 32B 2020-01-01T09:00:00 ... 2020-...
* lat (lat) float32 804B 33.5 33.51 33.52 ... 35.48 35.49 35.5
* lon (lon) float32 804B -75.5 -75.49 -75.48 ... -73.51 -73.5
Data variables:
analysed_sst (time, lat, lon) float64 1MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
analysis_error (time, lat, lon) float64 1MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
mask (time, lat, lon) float32 646kB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
sea_ice_fraction (time, lat, lon) float64 1MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
dt_1km_data (time, lat, lon) timedelta64[ns] 1MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
sst_anomaly (time, lat, lon) float64 1MB dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid Dimensions:
Coordinates: (3)
time
(time)
datetime64[ns]
2020-01-01T09:00:00 ... 2020-01-...
long_name : reference time of sst field standard_name : time axis : T comment : Nominal time of analyzed fields array(['2020-01-01T09:00:00.000000000', '2020-01-02T09:00:00.000000000',
'2020-01-03T09:00:00.000000000', '2020-01-04T09:00:00.000000000'],
dtype='datetime64[ns]') lat
(lat)
float32
33.5 33.51 33.52 ... 35.49 35.5
long_name : latitude standard_name : latitude axis : Y units : degrees_north valid_min : -90.0 valid_max : 90.0 comment : geolocations inherited from the input data without correction array([33.5 , 33.51, 33.52, ..., 35.48, 35.49, 35.5 ], dtype=float32) lon
(lon)
float32
-75.5 -75.49 ... -73.51 -73.5
long_name : longitude standard_name : longitude axis : X units : degrees_east valid_min : -180.0 valid_max : 180.0 comment : geolocations inherited from the input data without correction array([-75.5 , -75.49, -75.48, ..., -73.52, -73.51, -73.5 ], dtype=float32) Data variables: (6)
analysed_sst
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : analysed sea surface temperature standard_name : sea_surface_foundation_temperature units : kelvin valid_min : -32767 valid_max : 32767 comment : "Final" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
Bytes
1.23 MiB
315.63 kiB
Shape
(4, 201, 201)
(1, 201, 201)
Dask graph
4 chunks in 13 graph layers
Data type
float64 numpy.ndarray
201 201 4
analysis_error
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : estimated error standard deviation of analysed_sst units : kelvin valid_min : 0 valid_max : 32767 comment : uncertainty in "analysed_sst"
Bytes
1.23 MiB
315.63 kiB
Shape
(4, 201, 201)
(1, 201, 201)
Dask graph
4 chunks in 13 graph layers
Data type
float64 numpy.ndarray
201 201 4
mask
(time, lat, lon)
float32
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : sea/land field composite mask valid_min : 1 valid_max : 31 flag_masks : [ 1 2 4 8 16] flag_meanings : open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid comment : mask can be used to further filter the data. source : GMT "grdlandmask", ice flag from sea_ice_fraction data
Bytes
631.27 kiB
157.82 kiB
Shape
(4, 201, 201)
(1, 201, 201)
Dask graph
4 chunks in 13 graph layers
Data type
float32 numpy.ndarray
201 201 4
sea_ice_fraction
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : sea ice area fraction standard_name : sea_ice_area_fraction valid_min : 0 valid_max : 100 source : EUMETSAT OSI-SAF, copyright EUMETSAT comment : ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach.
Bytes
1.23 MiB
315.63 kiB
Shape
(4, 201, 201)
(1, 201, 201)
Dask graph
4 chunks in 13 graph layers
Data type
float64 numpy.ndarray
201 201 4
dt_1km_data
(time, lat, lon)
timedelta64[ns]
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : time to most recent 1km data valid_min : -127 valid_max : 127 source : MODIS and VIIRS pixels ingested by MUR comment : The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. "Fill value" indicates absence of such 1km data at the grid point.
Bytes
1.23 MiB
315.63 kiB
Shape
(4, 201, 201)
(1, 201, 201)
Dask graph
4 chunks in 13 graph layers
Data type
timedelta64[ns] numpy.ndarray
201 201 4
sst_anomaly
(time, lat, lon)
float64
dask.array<chunksize=(1, 201, 201), meta=np.ndarray>
long_name : SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period units : kelvin valid_min : -32767 valid_max : 32767 comment : anomaly reference to the day-of-year average between 2003 and 2014
Bytes
1.23 MiB
315.63 kiB
Shape
(4, 201, 201)
(1, 201, 201)
Dask graph
4 chunks in 13 graph layers
Data type
float64 numpy.ndarray
201 201 4
Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2020-01-01 09:00:00', '2020-01-02 09:00:00',
'2020-01-03 09:00:00', '2020-01-04 09:00:00'],
dtype='datetime64[ns]', name='time', freq=None)) PandasIndex
PandasIndex(Index([ 33.5, 33.5099983215332, 33.52000045776367,
33.529998779296875, 33.540000915527344, 33.54999923706055,
33.560001373291016, 33.56999969482422, 33.58000183105469,
33.59000015258789,
...
35.40999984741211, 35.41999816894531, 35.43000030517578,
35.439998626708984, 35.45000076293945, 35.459999084472656,
35.470001220703125, 35.47999954223633, 35.4900016784668,
35.5],
dtype='float32', name='lat', length=201)) PandasIndex
PandasIndex(Index([ -75.5, -75.48999786376953, -75.4800033569336,
-75.47000122070312, -75.45999908447266, -75.44999694824219,
-75.44000244140625, -75.43000030517578, -75.41999816894531,
-75.41000366210938,
...
-73.58999633789062, -73.58000183105469, -73.56999969482422,
-73.55999755859375, -73.55000305175781, -73.54000091552734,
-73.52999877929688, -73.5199966430664, -73.51000213623047,
-73.5],
dtype='float32', name='lon', length=201)) Attributes: (47)
Conventions : CF-1.7 title : Daily MUR SST, Final product summary : A merged, multi-sensor L4 Foundation SST analysis product from JPL. references : http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST institution : Jet Propulsion Laboratory history : created at nominal 4-day latency; replaced nrt (1-day latency) version. comment : MUR = "Multi-scale Ultra-high Resolution" license : These data are available free of charge under data policy of JPL PO.DAAC. id : MUR-JPL-L4-GLOB-v04.1 naming_authority : org.ghrsst product_version : 04.1 uuid : 27665bc0-d5fc-11e1-9b23-0800200c9a66 gds_version_id : 2.0 netcdf_version_id : 4.1 date_created : 20200124T180027Z start_time : 20200101T090000Z stop_time : 20200101T090000Z time_coverage_start : 20191231T210000Z time_coverage_end : 20200101T210000Z file_quality_level : 3 source : MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF platform : Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships sensor : MODIS, AMSR2, AVHRR, in-situ Metadata_Conventions : Unidata Observation Dataset v1.0 metadata_link : http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1 keywords : Oceans > Ocean Temperature > Sea Surface Temperature keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords standard_name_vocabulary : NetCDF Climate and Forecast (CF) Metadata Convention southernmost_latitude : -90.0 northernmost_latitude : 90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 spatial_resolution : 0.01 degrees geospatial_lat_units : degrees north geospatial_lat_resolution : 0.01 geospatial_lon_units : degrees east geospatial_lon_resolution : 0.01 acknowledgment : Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program. creator_name : JPL MUR SST project creator_email : ghrsst@podaac.jpl.nasa.gov creator_url : http://mur.jpl.nasa.gov project : NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program publisher_name : GHRSST Project Office publisher_url : http://www.ghrsst.org publisher_email : ghrsst-po@nceo.ac.uk processing_level : L4 cdm_data_type : grid
'analysed_sst' ].plot(x= 'lon' , y= 'lat' , col= "time" , col_wrap= 2 );
Summary
We learned how to subset xarray data cubes by time and space using sel() and slice(). Next we will show how to select via a shapefile. If you want to jump instead to creating monthly and seasonal means from a data cube, you can look at the 4-data-cube.ipynb tutorial or explore the gallery of xarray examples.