import warnings
from pathlib import Path
import echopype as ep # we recommend using "ep"
import xarray as xr
import hvplot.xarray # for interactive plots
import matplotlib.pyplot as pltIntro to echopype
Wu-Jung Lee (UW APL)
Getting started with Echopye
In this notebook, we will focus on the steps to convert instrument-generated raw data files, compute the volume backscattering strength (Sv) based on calibration and environmental parameters already stored in the file, and plot an echogram. We will use a .raw file generated by a Simrad EK80 echosounder as an example. Please see the Echopype documentation for usage with other supported formats.
Here we will use an example file collected as part of the Hake survey run by the NOAA Northwest Fisheries Science Center, stored in an open AWS data register from NOAA NCEI.
Echopype supports accessing data from multiple sources, such as a local filesystem (e.g., hard disk), a cloud object store (e.g., AWS S3), an http server, etc.
Here we directly access and convert data from an S3 bucket without having to download them first.
Convert raw data
Let’s take a look at what’s in ed, and EchoData object.
An EchoData object is a representation of an echopype-adapted version of the SONAR-netCDF4 version 1 convention. It shows the tree-like group structure in the netCDF data model. We aim for the conversion from raw data to the EchoData object as lossless as possible.
To see what we changed from the convention and why, please see the Data standardization section of the Echopype paper.
-
- conventions :
- CF-1.7, SONAR-netCDF4-1.0, ACDD-1.3
- keywords :
- EK80
- sonar_convention_authority :
- ICES
- sonar_convention_name :
- SONAR-netCDF4
- sonar_convention_version :
- 1.0
- summary :
- title :
- date_created :
- 2023-08-11T16:57:27Z
- processing_level :
- Level 1A
- processing_level_url :
- https://echopype.readthedocs.io/en/stable/processing-levels.html
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty* Attributes: conventions: CF-1.7, SONAR-netCDF4-1.0, ACDD-1.3 keywords: EK80 sonar_convention_authority: ICES sonar_convention_name: SONAR-netCDF4 sonar_convention_version: 1.0 summary: title: date_created: 2023-08-11T16:57:27Z processing_level: Level 1A processing_level_url: https://echopype.readthedocs.io/en/stable/pr...xarray.DatasetView -
- time1: 1
- sound_velocity_profile_depth: 2
- time1(time1)datetime64[ns]2023-08-11T16:57:27.277163
- axis :
- T
- long_name :
- Timestamps for NMEA position datagrams
- standard_name :
- time
- comment :
- Time coordinate corresponding to environmental variables. Note that Platform.time3 is the same as Environment.time1.
array(['2023-08-11T16:57:27.277163000'], dtype='datetime64[ns]')
- sound_velocity_profile_depth(sound_velocity_profile_depth)float641.0 1e+03
- standard_name :
- depth
- units :
- m
- axis :
- Z
- positive :
- down
- valid_min :
- 0.0
array([ 1., 1000.])
- depth(time1)float64200.0
array([200.])
- acidity(time1)float648.0
array([8.])
- salinity(time1)float6433.7
array([33.7])
- temperature(time1)float646.9
array([6.9])
- sound_speed_indicative(time1)float641.48e+03
array([1479.95204001])
- sound_velocity_profile(time1, sound_velocity_profile_depth)float641.48e+03 1.48e+03
- long_name :
- sound velocity profile
- standard_name :
- speed_of_sound_in_sea_water
- units :
- m/s
- valid_min :
- 0.0
- comment :
- parsed from raw data files as (depth, sound_speed) value pairs
array([[1479.95204, 1479.95204]])
- sound_velocity_source(time1)<U10'Calculated'
array(['Calculated'], dtype='<U10')
- transducer_name(time1)<U7'Unknown'
array(['Unknown'], dtype='<U7')
- transducer_sound_speed(time1)float641.48e+03
array([1480.])
- time1PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163'], dtype='datetime64[ns]', name='time1', freq=None))
- sound_velocity_profile_depthPandasIndex
PandasIndex(Index([1.0, 1000.0], dtype='float64', name='sound_velocity_profile_depth'))
<xarray.DatasetView> Size: 156B Dimensions: (time1: 1, sound_velocity_profile_depth: 2) Coordinates: * time1 (time1) datetime64[ns] 8B 2023-08-11T16:57:... * sound_velocity_profile_depth (sound_velocity_profile_depth) float64 16B ... Data variables: depth (time1) float64 8B 200.0 acidity (time1) float64 8B 8.0 salinity (time1) float64 8B 33.7 temperature (time1) float64 8B 6.9 sound_speed_indicative (time1) float64 8B 1.48e+03 sound_velocity_profile (time1, sound_velocity_profile_depth) float64 16B ... sound_velocity_source (time1) <U10 40B 'Calculated' transducer_name (time1) <U7 28B 'Unknown' transducer_sound_speed (time1) float64 8B 1.48e+03xarray.DatasetView -
- time1: 1451
- time2: 1837
- channel: 5
- time3: 1
- channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - time1(time1)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
- axis :
- T
- long_name :
- Timestamps for NMEA datagrams
- standard_name :
- time
- comment :
- Time coordinate corresponding to NMEA position data.
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', ..., '2023-08-11T17:05:30.095901000', '2023-08-11T17:05:29.858971000', '2023-08-11T17:05:30.086050000'], dtype='datetime64[ns]') - time2(time2)datetime64[ns]2023-08-11T16:57:28.465496 ... 2...
- axis :
- T
- long_name :
- Timestamps for platform motion and orientation data
- standard_name :
- time
- comment :
- Time coordinate corresponding to platform motion and orientation data.
array(['2023-08-11T16:57:28.465496000', '2023-08-11T16:57:28.739187000', '2023-08-11T16:57:28.953922000', ..., '2023-08-11T17:05:30.767675000', '2023-08-11T17:05:31.043379000', '2023-08-11T17:05:31.258127000'], dtype='datetime64[ns]') - time3(time3)datetime64[ns]2023-08-11T16:57:27.277163
- axis :
- T
- long_name :
- Timestamps for platform motion and orientation data from the Kongsberg Maritime Binary Datagram
- standard_name :
- time
- comment :
- Time coordinate corresponding to platform motion and orientation data from the Kongsberg Maritime Binary Datagram.
array(['2023-08-11T16:57:27.277163000'], dtype='datetime64[ns]')
- latitude(time1)float6443.95 43.95 43.95 ... 43.95 43.95
- long_name :
- Platform latitude
- standard_name :
- latitude
- units :
- degrees_north
- valid_range :
- (-90.0, 90.0)
array([43.94577833, 43.94577833, 43.94572517, ..., 43.9457045 , 43.94572 , 43.94572 ]) - longitude(time1)float64-125.0 -125.0 ... -124.9 -124.9
- long_name :
- Platform longitude
- standard_name :
- longitude
- units :
- degrees_east
- valid_range :
- (-180.0, 180.0)
array([-124.96901333, -124.96894833, -124.96879883, ..., -124.93656817, -124.93679333, -124.93679333]) - sentence_type(time1)<U3'GGA' 'GLL' 'GGA' ... 'GGA' 'GLL'
- long_name :
- NMEA sentence type
array(['GGA', 'GLL', 'GGA', ..., 'GGA', 'GGA', 'GLL'], dtype='<U3')
- pitch(time2)float64-0.01 -0.03 -0.04 ... 1.38 1.46
- long_name :
- Platform pitch
- standard_name :
- platform_pitch_angle
- units :
- arc_degree
- valid_range :
- (-90.0, 90.0)
array([-0.01 , -0.03 , -0.04 , ..., 1.25999999, 1.38 , 1.46000004]) - roll(time2)float64-0.11 0.26 0.51 ... 0.86 0.87 0.86
- long_name :
- Platform roll
- standard_name :
- platform_roll_angle
- units :
- arc_degree
- valid_range :
- (-90.0, 90.0)
array([-0.11 , 0.25999999, 0.50999999, ..., 0.86000001, 0.87 , 0.86000001]) - vertical_offset(time2)float640.71 0.64 0.58 ... -0.61 -0.62
- long_name :
- Platform vertical offset from nominal water level
- units :
- m
array([ 0.70999998, 0.63999999, 0.57999998, ..., -0.58999997, -0.61000001, -0.62 ]) - water_level()float640.0
- long_name :
- Distance from the platform coordinate system origin to the nominal water level along the z-axis
- units :
- m
array(0.)
- drop_keel_offset()float649.15
array(9.15)
- drop_keel_offset_is_manual()int641
array(1)
- water_level_draft_is_manual()int640
array(0)
- transducer_offset_x(channel)float640.0 0.0 0.0 0.0 0.0
- long_name :
- x-axis distance from the platform coordinate system origin to the sonar transducer
- units :
- m
array([0., 0., 0., 0., 0.])
- transducer_offset_y(channel)float640.0 0.0 0.0 0.0 0.0
- long_name :
- y-axis distance from the platform coordinate system origin to the sonar transducer
- units :
- m
array([0., 0., 0., 0., 0.])
- transducer_offset_z(channel)float640.0 0.0 0.0 0.0 0.0
- long_name :
- z-axis distance from the platform coordinate system origin to the sonar transducer
- units :
- m
array([0., 0., 0., 0., 0.])
- MRU_offset_x()float64nan
- long_name :
- Distance along the x-axis from the platform coordinate system origin to the motion reference unit sensor origin
- units :
- m
array(nan)
- MRU_offset_y()float64nan
- long_name :
- Distance along the y-axis from the platform coordinate system origin to the motion reference unit sensor origin
- units :
- m
array(nan)
- MRU_offset_z()float64nan
- long_name :
- Distance along the z-axis from the platform coordinate system origin to the motion reference unit sensor origin
- units :
- m
array(nan)
- MRU_rotation_x()float64nan
- long_name :
- Extrinsic rotation about the x-axis from the platform to MRU coordinate systems
- units :
- arc_degree
- valid_range :
- (–180.0, 180.0)
array(nan)
- MRU_rotation_y()float64nan
- long_name :
- Extrinsic rotation about the y-axis from the platform to MRU coordinate systems
- units :
- arc_degree
- valid_range :
- (–180.0, 180.0)
array(nan)
- MRU_rotation_z()float64nan
- long_name :
- Extrinsic rotation about the z-axis from the platform to MRU coordinate systems
- units :
- arc_degree
- valid_range :
- (–180.0, 180.0)
array(nan)
- position_offset_x()float64nan
- long_name :
- Distance along the x-axis from the platform coordinate system origin to the latitude/longitude sensor origin
- units :
- m
array(nan)
- position_offset_y()float64nan
- long_name :
- Distance along the y-axis from the platform coordinate system origin to the latitude/longitude sensor origin
- units :
- m
array(nan)
- position_offset_z()float64nan
- long_name :
- Distance along the z-axis from the platform coordinate system origin to the latitude/longitude sensor origin
- units :
- m
array(nan)
- frequency_nominal(channel)float641.2e+05 1.8e+04 7e+04 3.8e+04 2e+05
- units :
- Hz
- long_name :
- Transducer frequency
- valid_min :
- 0.0
- standard_name :
- sound_frequency
array([120000., 18000., 70000., 38000., 200000.])
- heading(time2)float6494.5 94.5 94.5 ... 91.4 91.4 91.4
- long_name :
- Platform heading (true)
- standard_name :
- platform_orientation
- units :
- degrees_north
- valid_min :
- 0.0
- valid_max :
- 360.0
array([94.5 , 94.5 , 94.5 , ..., 91.40000153, 91.40000153, 91.40000153]) - latitude_mru1(time3)float64nan
- long_name :
- Platform latitude
- standard_name :
- latitude
- units :
- degrees_north
- valid_range :
- (-90.0, 90.0)
- comment :
- Derived from the Simrad MRU1 Datagrams which are a wrapper of the Kongsberg Maritime Binary Datagrams.
array([nan])
- longitude_mru1(time3)float64nan
- long_name :
- Platform longitude
- standard_name :
- longitude
- units :
- degrees_east
- valid_range :
- (-180.0, 180.0)
- comment :
- Derived from the Simrad MRU1 Datagrams which are a wrapper of the Kongsberg Maritime Binary Datagrams.
array([nan])
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - time1PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:28.084934', '2023-08-11 16:57:28.313240', '2023-08-11 16:57:28.917156', '2023-08-11 16:57:29.089186', '2023-08-11 16:57:29.349422', '2023-08-11 16:57:30.080532', '2023-08-11 16:57:29.869759', ... '2023-08-11 17:05:27.311052', '2023-08-11 17:05:28.082108', '2023-08-11 17:05:27.919565', '2023-08-11 17:05:28.302895', '2023-08-11 17:05:28.919241', '2023-08-11 17:05:29.088322', '2023-08-11 17:05:29.324667', '2023-08-11 17:05:30.095901', '2023-08-11 17:05:29.858971', '2023-08-11 17:05:30.086050'], dtype='datetime64[ns]', name='time1', length=1451, freq=None)) - time2PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:28.465496', '2023-08-11 16:57:28.739187', '2023-08-11 16:57:28.953922', '2023-08-11 16:57:29.260298', '2023-08-11 16:57:29.568590', '2023-08-11 16:57:29.844364', '2023-08-11 16:57:30.059101', '2023-08-11 16:57:30.366383', '2023-08-11 16:57:30.642157', '2023-08-11 16:57:30.856892', ... '2023-08-11 17:05:28.957245', '2023-08-11 17:05:29.173046', '2023-08-11 17:05:29.448530', '2023-08-11 17:05:29.665287', '2023-08-11 17:05:29.970560', '2023-08-11 17:05:30.246332', '2023-08-11 17:05:30.461327', '2023-08-11 17:05:30.767675', '2023-08-11 17:05:31.043379', '2023-08-11 17:05:31.258127'], dtype='datetime64[ns]', name='time2', length=1837, freq=None)) - time3PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163'], dtype='datetime64[ns]', name='time3', freq=None))
- platform_name :
- platform_type :
- platform_code_ICES :
<xarray.DatasetView> Size: 127kB Dimensions: (time1: 1451, time2: 1837, channel: 5, time3: 1) Coordinates: * channel (channel) <U25 500B 'WBT 400140-15 ES120-7C_... * time1 (time1) datetime64[ns] 12kB 2023-08-11T16:57... * time2 (time2) datetime64[ns] 15kB 2023-08-11T16:57... * time3 (time3) datetime64[ns] 8B 2023-08-11T16:57:2... Data variables: (12/26) latitude (time1) float64 12kB 43.95 43.95 ... 43.95 longitude (time1) float64 12kB -125.0 -125.0 ... -124.9 sentence_type (time1) <U3 17kB 'GGA' 'GLL' ... 'GGA' 'GLL' pitch (time2) float64 15kB -0.01 -0.03 ... 1.38 1.46 roll (time2) float64 15kB -0.11 0.26 ... 0.87 0.86 vertical_offset (time2) float64 15kB 0.71 0.64 ... -0.61 -0.62 ... ... position_offset_y float64 8B nan position_offset_z float64 8B nan frequency_nominal (channel) float64 40B 1.2e+05 1.8e+04 ... 2e+05 heading (time2) float64 15kB 94.5 94.5 ... 91.4 91.4 latitude_mru1 (time3) float64 8B nan longitude_mru1 (time3) float64 8B nan Attributes: platform_name: platform_type: platform_code_ICES:xarray.DatasetView -
- nmea_time: 5479
- channel: 5
- time1: 1451
- time2: 1837
- time3: 1
- channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - time1(time1)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
- axis :
- T
- long_name :
- Timestamps for NMEA datagrams
- standard_name :
- time
- comment :
- Time coordinate corresponding to NMEA position data.
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', ..., '2023-08-11T17:05:30.095901000', '2023-08-11T17:05:29.858971000', '2023-08-11T17:05:30.086050000'], dtype='datetime64[ns]') - time2(time2)datetime64[ns]2023-08-11T16:57:28.465496 ... 2...
- axis :
- T
- long_name :
- Timestamps for platform motion and orientation data
- standard_name :
- time
- comment :
- Time coordinate corresponding to platform motion and orientation data.
array(['2023-08-11T16:57:28.465496000', '2023-08-11T16:57:28.739187000', '2023-08-11T16:57:28.953922000', ..., '2023-08-11T17:05:30.767675000', '2023-08-11T17:05:31.043379000', '2023-08-11T17:05:31.258127000'], dtype='datetime64[ns]') - time3(time3)datetime64[ns]2023-08-11T16:57:27.277163
- axis :
- T
- long_name :
- Timestamps for platform motion and orientation data from the Kongsberg Maritime Binary Datagram
- standard_name :
- time
- comment :
- Time coordinate corresponding to platform motion and orientation data from the Kongsberg Maritime Binary Datagram.
array(['2023-08-11T16:57:27.277163000'], dtype='datetime64[ns]')
- nmea_time(nmea_time)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
- axis :
- T
- long_name :
- Timestamps for NMEA datagrams
- standard_name :
- time
- comment :
- Time coordinate corresponding to NMEA sensor data.
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', ..., '2023-08-11T17:05:30.086050000', '2023-08-11T17:05:30.186049000', '2023-08-11T17:05:30.261047000'], dtype='datetime64[ns]')
- NMEA_datagram(nmea_time)<U70'$SDVLW,4418.987,N,4418.987,N' ....
- long_name :
- NMEA datagram
array(['$SDVLW,4418.987,N,4418.987,N', '$GPALR,,,V,V,', '$GPDTM,W84,,00.0000,N,00.0000,E,,W84', ..., '$GPGLL,4356.7432,N,12456.2076,W,170529.00,A,D', '$GPZDA,170530.00,11,08,2023,00,00', '$GPVTG,89.8,T,74.9,M,10.357,N,19.181,K,D'], dtype='<U70')
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - time1PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:28.084934', '2023-08-11 16:57:28.313240', '2023-08-11 16:57:28.917156', '2023-08-11 16:57:29.089186', '2023-08-11 16:57:29.349422', '2023-08-11 16:57:30.080532', '2023-08-11 16:57:29.869759', ... '2023-08-11 17:05:27.311052', '2023-08-11 17:05:28.082108', '2023-08-11 17:05:27.919565', '2023-08-11 17:05:28.302895', '2023-08-11 17:05:28.919241', '2023-08-11 17:05:29.088322', '2023-08-11 17:05:29.324667', '2023-08-11 17:05:30.095901', '2023-08-11 17:05:29.858971', '2023-08-11 17:05:30.086050'], dtype='datetime64[ns]', name='time1', length=1451, freq=None)) - time2PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:28.465496', '2023-08-11 16:57:28.739187', '2023-08-11 16:57:28.953922', '2023-08-11 16:57:29.260298', '2023-08-11 16:57:29.568590', '2023-08-11 16:57:29.844364', '2023-08-11 16:57:30.059101', '2023-08-11 16:57:30.366383', '2023-08-11 16:57:30.642157', '2023-08-11 16:57:30.856892', ... '2023-08-11 17:05:28.957245', '2023-08-11 17:05:29.173046', '2023-08-11 17:05:29.448530', '2023-08-11 17:05:29.665287', '2023-08-11 17:05:29.970560', '2023-08-11 17:05:30.246332', '2023-08-11 17:05:30.461327', '2023-08-11 17:05:30.767675', '2023-08-11 17:05:31.043379', '2023-08-11 17:05:31.258127'], dtype='datetime64[ns]', name='time2', length=1837, freq=None)) - time3PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163'], dtype='datetime64[ns]', name='time3', freq=None))
- nmea_timePandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', ... '2023-08-11 17:05:29.960487', '2023-08-11 17:05:30.095901', '2023-08-11 17:05:30.247984', '2023-08-11 17:05:30.281317', '2023-08-11 17:05:29.777723', '2023-08-11 17:05:29.858971', '2023-08-11 17:05:30.004803', '2023-08-11 17:05:30.086050', '2023-08-11 17:05:30.186049', '2023-08-11 17:05:30.261047'], dtype='datetime64[ns]', name='nmea_time', length=5479, freq=None))
- description :
- All NMEA sensor datagrams
<xarray.DatasetView> Size: 2MB Dimensions: (nmea_time: 5479, channel: 5, time1: 1451, time2: 1837, time3: 1) Coordinates: * channel (channel) <U25 500B 'WBT 400140-15 ES120-7C_ES' ... 'WBT 4... * time1 (time1) datetime64[ns] 12kB 2023-08-11T16:57:27.277163 ...... * time2 (time2) datetime64[ns] 15kB 2023-08-11T16:57:28.465496 ...... * time3 (time3) datetime64[ns] 8B 2023-08-11T16:57:27.277163 * nmea_time (nmea_time) datetime64[ns] 44kB 2023-08-11T16:57:27.277163... Data variables: NMEA_datagram (nmea_time) <U70 2MB '$SDVLW,4418.987,N,4418.987,N' ... '$... Attributes: description: All NMEA sensor datagramsxarray.DatasetView -
- filenames: 1
- filenames(filenames)int640
- long_name :
- Index for data and metadata source filenames
array([0])
- source_filenames(filenames)<U82's3://noaa-wcsd-pds/data/raw/Bel...
- long_name :
- Source filenames
array(['s3://noaa-wcsd-pds/data/raw/Bell_M._Shimada/SH2306/EK80/Hake-D20230811-T165727.raw'], dtype='<U82')
- filenamesPandasIndex
PandasIndex(Index([0], dtype='int64', name='filenames'))
- conversion_software_name :
- echopype
- conversion_software_version :
- 0.10.1
- conversion_time :
- 2025-04-25T18:18:28Z
<xarray.DatasetView> Size: 336B Dimensions: (filenames: 1) Coordinates: * filenames (filenames) int64 8B 0 Data variables: source_filenames (filenames) <U82 328B 's3://noaa-wcsd-pds/data/raw/Bell... Attributes: conversion_software_name: echopype conversion_software_version: 0.10.1 conversion_time: 2025-04-25T18:18:28Zxarray.DatasetView -
- channel_all: 5
- beam_group: 1
- channel_all(channel_all)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - beam_group(beam_group)<U11'Beam_group1'
- long_name :
- Beam group name
array(['Beam_group1'], dtype='<U11')
- frequency_nominal(channel_all)float641.2e+05 1.8e+04 7e+04 3.8e+04 2e+05
- units :
- Hz
- long_name :
- Transducer frequency
- valid_min :
- 0.0
- standard_name :
- sound_frequency
array([120000., 18000., 70000., 38000., 200000.])
- transceiver_serial_number(channel_all)<U6'400140' '400141' ... '400145'
- long_name :
- Transceiver serial number
array(['400140', '400141', '400142', '400143', '400145'], dtype='<U6')
- transducer_name(channel_all)<U4'ES18' 'ES18' 'ES18' 'ES18' 'ES18'
- long_name :
- Transducer name
array(['ES18', 'ES18', 'ES18', 'ES18', 'ES18'], dtype='<U4')
- transducer_serial_number(channel_all)<U1'0' '0' '0' '0' '0'
- long_name :
- Transducer serial number
array(['0', '0', '0', '0', '0'], dtype='<U1')
- beam_group_descr(beam_group)<U131'contains backscatter power (unc...
- long_name :
- Beam group description
array(['contains backscatter power (uncalibrated) and other beam or channel-specific data, including split-beam angle data when they exist.'], dtype='<U131')
- channel_allPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel_all')) - beam_groupPandasIndex
PandasIndex(Index(['Beam_group1'], dtype='object', name='beam_group'))
- sonar_manufacturer :
- Simrad
- sonar_model :
- EK80
- sonar_serial_number :
- sonar_software_name :
- EK80
- sonar_software_version :
- 21.15.2.0
- sonar_type :
- echosounder
<xarray.DatasetView> Size: 1kB Dimensions: (channel_all: 5, beam_group: 1) Coordinates: * channel_all (channel_all) <U25 500B 'WBT 400140-15 ES120-7... * beam_group (beam_group) <U11 44B 'Beam_group1' Data variables: frequency_nominal (channel_all) float64 40B 1.2e+05 ... 2e+05 transceiver_serial_number (channel_all) <U6 120B '400140' ... '400145' transducer_name (channel_all) <U4 80B 'ES18' 'ES18' ... 'ES18' transducer_serial_number (channel_all) <U1 20B '0' '0' '0' '0' '0' beam_group_descr (beam_group) <U131 524B 'contains backscatter ... Attributes: sonar_manufacturer: Simrad sonar_model: EK80 sonar_serial_number: sonar_software_name: EK80 sonar_software_version: 21.15.2.0 sonar_type: echosounderxarray.DatasetView -
- channel: 5
- ping_time: 213
- range_sample: 36198
- transmit_sample: 148
- channel_all: 5
- beam_group: 1
- channel_all(channel_all)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - beam_group(beam_group)<U11'Beam_group1'
- long_name :
- Beam group name
array(['Beam_group1'], dtype='<U11')
- channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - ping_time(ping_time)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
- long_name :
- Timestamp of each ping
- standard_name :
- time
- axis :
- T
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:30.276106000', '2023-08-11T16:57:33.288034000', ..., '2023-08-11T17:05:25.278637000', '2023-08-11T17:05:27.281294000', '2023-08-11T17:05:29.279934000'], dtype='datetime64[ns]') - range_sample(range_sample)int640 1 2 3 ... 36194 36195 36196 36197
- long_name :
- Along-range sample number, base 0
array([ 0, 1, 2, ..., 36195, 36196, 36197])
- transmit_sample(transmit_sample)int640 1 2 3 4 5 ... 143 144 145 146 147
- long_name :
- Transmit pulse sample number, base 0
- comment :
- Only exist for Simrad EK80 file with RAW4 datagrams
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147])
- frequency_nominal(channel)float641.2e+05 1.8e+04 7e+04 3.8e+04 2e+05
- units :
- Hz
- long_name :
- Transducer frequency
- valid_min :
- 0.0
- standard_name :
- sound_frequency
array([120000., 18000., 70000., 38000., 200000.])
- beam_type(channel)int641 1 1 1 1
- long_name :
- type of transducer (0-single, 1-split)
array([1, 1, 1, 1, 1])
- beamwidth_twoway_alongship(channel)float646.63 10.92 6.55 6.71 6.79
- long_name :
- Half power two-way beam width along alongship axis of beam
- units :
- arc_degree
- valid_range :
- (0.0, 360.0)
- comment :
- Introduced in echopype for Simrad echosounders to avoid potential confusion with convention definitions. The alongship angle corresponds to the minor angle in SONAR-netCDF4 vers 2. The convention defines one-way transmit or receive beamwidth (beamwidth_receive_minor and beamwidth_transmit_minor), but Simrad echosounders record two-way beamwidth in the data.
array([ 6.63, 10.92, 6.55, 6.71, 6.79])
- beamwidth_twoway_athwartship(channel)float646.74 10.6 6.61 6.81 6.07
- long_name :
- Half power two-way beam width along athwartship axis of beam
- units :
- arc_degree
- valid_range :
- (0.0, 360.0)
- comment :
- Introduced in echopype for Simrad echosounders to avoid potential confusion with convention definitions. The athwartship angle corresponds to the major angle in SONAR-netCDF4 vers 2. The convention defines one-way transmit or receive beamwidth (beamwidth_receive_major and beamwidth_transmit_major), but Simrad echosounders record two-way beamwidth in the data.
array([ 6.74, 10.6 , 6.61, 6.81, 6.07])
- beam_direction_x(channel)float64nan nan nan nan nan
- long_name :
- x-component of the vector that gives the pointing direction of the beam, in sonar beam coordinate system
- units :
- 1
- valid_range :
- (-1.0, 1.0)
array([nan, nan, nan, nan, nan])
- beam_direction_y(channel)float64nan nan nan nan nan
- long_name :
- y-component of the vector that gives the pointing direction of the beam, in sonar beam coordinate system
- units :
- 1
- valid_range :
- (-1.0, 1.0)
array([nan, nan, nan, nan, nan])
- beam_direction_z(channel)float64nan nan nan nan nan
- long_name :
- z-component of the vector that gives the pointing direction of the beam, in sonar beam coordinate system
- units :
- 1
- valid_range :
- (-1.0, 1.0)
array([nan, nan, nan, nan, nan])
- angle_offset_alongship(channel)float64-0.04 -0.11 -0.02 -0.2 -0.07
- long_name :
- electrical alongship angle offset of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The alongship angle corresponds to the minor angle in SONAR-netCDF4 vers 2.
array([-0.04, -0.11, -0.02, -0.2 , -0.07])
- angle_offset_athwartship(channel)float640.17 -0.05 0.0 -0.17 0.25
- long_name :
- electrical athwartship angle offset of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The athwartship angle corresponds to the major angle in SONAR-netCDF4 vers 2.
array([ 0.17, -0.05, 0. , -0.17, 0.25])
- angle_sensitivity_alongship(channel)float6423.0 15.5 23.0 23.0 23.0
- long_name :
- alongship angle sensitivity of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The alongship angle corresponds to the minor angle in SONAR-netCDF4 vers 2.
array([23. , 15.5, 23. , 23. , 23. ])
- angle_sensitivity_athwartship(channel)float6423.0 15.5 23.0 23.0 23.0
- long_name :
- athwartship angle sensitivity of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The athwartship angle corresponds to the major angle in SONAR-netCDF4 vers 2.
array([23. , 15.5, 23. , 23. , 23. ])
- equivalent_beam_angle(channel)float64-20.7 -17.0 -20.7 -20.7 -20.7
- long_name :
- Equivalent beam angle
- units :
- sr
- valid_range :
- (0.0, 12.566370614359172)
array([-20.7, -17. , -20.7, -20.7, -20.7])
- transceiver_software_version(channel)<U4'2.54' '2.54' '2.54' '2.54' '2.54'
array(['2.54', '2.54', '2.54', '2.54', '2.54'], dtype='<U4')
- beam_stabilisation()int80
- long_name :
- Beam stabilisation applied (or not)
- flag_values :
- [0, 1]
- flag_meanings :
- ['not stabilised', 'stabilised']
array(0, dtype=int8)
- non_quantitative_processing()int160
- long_name :
- Presence or not of non-quantitative processing applied to the backscattering data (sonar specific)
- flag_values :
- [0]
- flag_meanings :
- ['None']
array(0, dtype=int16)
- transmit_frequency_start(channel)float641.2e+05 1.8e+04 7e+04 3.8e+04 2e+05
- long_name :
- Start frequency in transmitted pulse
- units :
- Hz
- standard_name :
- sound_frequency
- valid_min :
- 0.0
array([120000., 18000., 70000., 38000., 200000.])
- transmit_frequency_stop(channel)float641.2e+05 1.8e+04 7e+04 3.8e+04 2e+05
- long_name :
- Stop frequency in transmitted pulse
- units :
- Hz
- standard_name :
- sound_frequency
- valid_min :
- 0.0
array([120000., 18000., 70000., 38000., 200000.])
- backscatter_r(channel, ping_time, range_sample)float3210.38 13.88 16.02 ... nan nan nan
- long_name :
- Raw backscatter measurements (real part)
- units :
- dB
array([[[ 10.383183 , 13.875601 , 16.015736 , ..., nan, nan, nan], [ 10.430219 , 13.910878 , 16.051014 , ..., nan, nan, nan], [ 10.383183 , 13.875601 , 16.015736 , ..., nan, nan, nan], ..., [ 10.465496 , 13.969673 , 16.121567 , ..., nan, nan, nan], [ 10.465496 , 13.969673 , 16.121567 , ..., nan, nan, nan], [ 10.430219 , 13.899119 , 16.051014 , ..., nan, nan, nan]], [[ -11.782502 , -5.7031074 , -0.50563633, ..., -125.868164 , -126.09158 , -126.29149 ], [ -11.711948 , -5.6443124 , -0.47035936, ..., -133.06467 , -130.23074 , -127.82016 ], [ -11.770743 , -5.6795893 , -0.49387732, ..., -133.14697 , -132.48848 , -131.8182 ], ... [ 3.5394542 , 10.41846 , 15.780557 , ..., nan, nan, nan], [ 3.4689002 , 10.383183 , 15.757039 , ..., nan, nan, nan], [ 3.4924183 , 10.383183 , 15.757039 , ..., nan, nan, nan]], [[ 6.067636 , 8.866274 , 10.747711 , ..., nan, nan, nan], [ 6.079395 , 8.842756 , 10.747711 , ..., nan, nan, nan], [ 6.2087436 , 9.019141 , 10.912337 , ..., nan, nan, nan], ..., [ 6.3028154 , 9.077936 , 10.959373 , ..., nan, nan, nan], [ 6.2087436 , 9.0309 , 10.935855 , ..., nan, nan, nan], [ 6.3028154 , 9.077936 , 10.971132 , ..., nan, nan, nan]]], dtype=float32) - transmit_pulse_r(channel, ping_time, transmit_sample)float6430.66 30.91 33.56 ... nan nan nan
- long_name :
- Real part of the transmit pulse
- units :
- V
- comment :
- Only exist for Simrad EK80 file with RAW4 datagrams
array([[[ 30.65556717, 30.90556717, 33.56114578, ..., nan, nan, nan], [ 34.56123352, 34.81124496, 37.56124878, ..., nan, nan, nan], [ 30.65556908, 30.90556717, 33.56114578, ..., nan, nan, nan], ..., [ 37.06131744, 37.5613327 , 40.56132889, ..., nan, nan, nan], [ 37.06131744, 37.5613327 , 40.5613327 , ..., nan, nan, nan], [ 34.56123352, 34.81124496, 37.56124878, ..., nan, nan, nan]], [[ -2.27714586, -2.35527086, -2.35527182, ..., -0.14331225, -0.16089043, -0.14624244], [ -2.24589515, -2.27714658, -2.30839705, ..., -0.1345232 , -0.15112484, -0.13647681], [ -2.24589539, -2.32402134, -2.33964634, ..., -0.14331223, -0.16089046, -0.14526588], ... [-16.59304237, -17.46806908, -17.84306908, ..., nan, nan, nan], [-16.09304428, -16.96807098, -17.46807098, ..., nan, nan, nan], [-16.21804619, -17.09307098, -17.46807098, ..., nan, nan, nan]], [[-17.46784592, -16.46785927, -17.84284973, ..., nan, nan, nan], [-17.46784592, -16.46786118, -17.84284973, ..., nan, nan, nan], [ -5.96050882, -5.17925167, -5.8980217 , ..., nan, nan, nan], ..., [-21.96796036, -21.09298134, -22.46796227, ..., nan, nan, nan], [ -5.96051025, -5.17925501, -5.89802504, ..., nan, nan, nan], [-21.96796036, -21.09298134, -22.46796227, ..., nan, nan, nan]]]) - transmit_pulse_i(channel, ping_time, transmit_sample)float64-23.34 -23.22 -26.34 ... nan nan
- long_name :
- Imaginary part of the transmit pulse
- units :
- V
- comment :
- Only exist for Simrad EK80 file with RAW4 datagrams
array([[[-23.34309959, -23.21811867, -26.34310722, ..., nan, nan, nan], [-17.84304619, -17.46806526, -20.46805382, ..., nan, nan, nan], [-23.34309959, -23.21811867, -26.34310722, ..., nan, nan, nan], ..., [-12.04647541, -11.67149448, -13.73398113, ..., nan, nan, nan], [-12.04647541, -11.67149448, -13.73398113, ..., nan, nan, nan], [-17.84304619, -17.46806526, -20.46805382, ..., nan, nan, nan]], [[ -2.05839658, -2.15214849, -2.15214658, ..., -0.34911257, -0.36668324, -0.3930521 ], [ -2.12089944, -2.26152492, -2.24589849, ..., -0.35106641, -0.36863816, -0.39695752], [ -2.08964658, -2.19902253, -2.12089705, ..., -0.34911197, -0.36668444, -0.39500636], ... [ 4.61689329, 5.33564615, 5.14814711, ..., nan, nan, nan], [ 5.49187517, 6.27312899, 5.99187899, ..., nan, nan, nan], [ 5.49187517, 6.27312803, 5.92937899, ..., nan, nan, nan]], [[ 16.90547752, 16.65550423, 17.53047371, ..., nan, nan, nan], [ 16.90547752, 16.78050423, 17.53046989, ..., nan, nan, nan], [ 23.90554619, 23.40556145, 24.65554619, ..., nan, nan, nan], ..., [ 11.70268726, 11.95269871, 12.26518536, ..., nan, nan, nan], [ 23.90554619, 23.40556145, 24.65554619, ..., nan, nan, nan], [ 11.70268917, 11.95269871, 12.26518536, ..., nan, nan, nan]]]) - angle_athwartship(channel, ping_time, range_sample)float320.0 0.0 0.0 0.0 ... nan nan nan nan
- long_name :
- electrical athwartship angle
- comment :
- Introduced in echopype for Simrad echosounders. The athwartship angle corresponds to the major angle in SONAR-netCDF4 vers 2.
array([[[ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], ..., [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan]], [[ 0., 0., 0., ..., 21., 5., -13.], [ -1., 0., 0., ..., 52., 49., 45.], [ 0., 0., 0., ..., -81., -87., -90.], ..., [ 0., 0., 0., ..., -51., -57., -61.], [ 0., 0., 0., ..., -18., -16., -16.], [ -1., 0., 0., ..., 42., 38., 34.]], [[ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], ..., [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan]], [[ 1., 1., 1., ..., nan, nan, nan], [ 1., 1., 1., ..., nan, nan, nan], [ 1., 1., 1., ..., nan, nan, nan], ..., [ 1., 1., 1., ..., nan, nan, nan], [ 1., 1., 1., ..., nan, nan, nan], [ 1., 1., 1., ..., nan, nan, nan]], [[ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], ..., [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan]]], dtype=float32) - angle_alongship(channel, ping_time, range_sample)float32-1.0 -1.0 -1.0 -1.0 ... nan nan nan
- long_name :
- electrical alongship angle
- comment :
- Introduced in echopype for Simrad echosounders. The alongship angle corresponds to the minor angle in SONAR-netCDF4 vers 2.
array([[[ -1., -1., -1., ..., nan, nan, nan], [ -1., -1., -1., ..., nan, nan, nan], [ -1., -1., -1., ..., nan, nan, nan], ..., [ -1., -1., -1., ..., nan, nan, nan], [ -1., -1., -1., ..., nan, nan, nan], [ -1., -1., -1., ..., nan, nan, nan]], [[ 0., 0., 0., ..., 28., 31., 28.], [ -1., 0., 0., ..., -25., -40., -48.], [ -1., 0., 0., ..., -26., -37., -49.], ..., [ 0., 0., 0., ..., 83., 81., 83.], [ -1., 0., 0., ..., -34., -39., -47.], [ -1., 0., 0., ..., -6., -10., -11.]], [[ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], ..., [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan]], [[ 1., 0., 0., ..., nan, nan, nan], [ 1., 1., 0., ..., nan, nan, nan], [ 1., 0., 0., ..., nan, nan, nan], ..., [ 0., 0., 0., ..., nan, nan, nan], [ 1., 1., 0., ..., nan, nan, nan], [ 1., 1., 0., ..., nan, nan, nan]], [[ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], [ 0., 0., 0., ..., nan, nan, nan], ..., [ 0., 0., -1., ..., nan, nan, nan], [ -1., 0., -1., ..., nan, nan, nan], [ 0., 0., -1., ..., nan, nan, nan]]], dtype=float32) - sample_interval(channel, ping_time)float644e-05 4e-05 ... 3.2e-05 3.2e-05
- long_name :
- Interval between recorded raw data samples
- units :
- s
- valid_min :
- 0.0
array([[4.0e-05, 4.0e-05, 4.0e-05, ..., 4.0e-05, 4.0e-05, 4.0e-05], [2.8e-05, 2.8e-05, 2.8e-05, ..., 2.8e-05, 2.8e-05, 2.8e-05], [4.8e-05, 4.8e-05, 4.8e-05, ..., 4.8e-05, 4.8e-05, 4.8e-05], [4.0e-05, 4.0e-05, 4.0e-05, ..., 4.0e-05, 4.0e-05, 4.0e-05], [3.2e-05, 3.2e-05, 3.2e-05, ..., 3.2e-05, 3.2e-05, 3.2e-05]]) - transmit_power(channel, ping_time)float64250.0 250.0 250.0 ... 105.0 105.0
- long_name :
- Nominal transmit power
- units :
- W
- valid_min :
- 0.0
array([[ 250., 250., 250., ..., 250., 250., 250.], [1000., 1000., 1000., ..., 1000., 1000., 1000.], [ 750., 750., 750., ..., 750., 750., 750.], [2000., 2000., 2000., ..., 2000., 2000., 2000.], [ 105., 105., 105., ..., 105., 105., 105.]]) - transmit_duration_nominal(channel, ping_time)float320.001024 0.001024 ... 0.001024
- long_name :
- Nominal bandwidth of transmitted pulse
- units :
- s
- valid_min :
- 0.0
array([[0.001024, 0.001024, 0.001024, ..., 0.001024, 0.001024, 0.001024], [0.001024, 0.001024, 0.001024, ..., 0.001024, 0.001024, 0.001024], [0.001024, 0.001024, 0.001024, ..., 0.001024, 0.001024, 0.001024], [0.001024, 0.001024, 0.001024, ..., 0.001024, 0.001024, 0.001024], [0.001024, 0.001024, 0.001024, ..., 0.001024, 0.001024, 0.001024]], dtype=float32) - slope(channel, ping_time)float640.01628 0.01628 ... 0.009766
- long_name :
- Hann window slope parameter for transmit signal
array([[0.016276 , 0.016276 , 0.016276 , ..., 0.016276 , 0.016276 , 0.016276 ], [0.5 , 0.5 , 0.5 , ..., 0.5 , 0.5 , 0.5 ], [0.0279018 , 0.0279018 , 0.0279018 , ..., 0.0279018 , 0.0279018 , 0.0279018 ], [0.25699 , 0.25699 , 0.25699 , ..., 0.25699 , 0.25699 , 0.25699 ], [0.00976563, 0.00976563, 0.00976563, ..., 0.00976563, 0.00976563, 0.00976563]]) - channel_mode(channel, ping_time)int80 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 0
- long_name :
- Transceiver mode
- flag_values :
- [0, 1]
- flag_meanings :
- ['Active', 'Unknown']
array([[0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0]], dtype=int8) - transmit_type(channel, ping_time)<U3'CW' 'CW' 'CW' ... 'CW' 'CW' 'CW'
- long_name :
- Type of transmitted pulse
- flag_values :
- ['CW', 'LFM', 'FMD']
- flag_meanings :
- ['Continuous Wave – a pulse nominally of one frequency', 'Linear Frequency Modulation – a pulse which varies from transmit_frequency_start to transmit_frequency_stop in a linear manner over the nominal pulse duration (transmit_duration_nominal)', "Frequency Modulated 'D' - An EK80-specific FM type that is not clearly described"]
array([['CW', 'CW', 'CW', ..., 'CW', 'CW', 'CW'], ['CW', 'CW', 'CW', ..., 'CW', 'CW', 'CW'], ['CW', 'CW', 'CW', ..., 'CW', 'CW', 'CW'], ['CW', 'CW', 'CW', ..., 'CW', 'CW', 'CW'], ['CW', 'CW', 'CW', ..., 'CW', 'CW', 'CW']], dtype='<U3') - sample_time_offset(channel, ping_time)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- Time offset that is subtracted from the timestamp of each sample
- units :
- s
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]])
- channel_allPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel_all')) - beam_groupPandasIndex
PandasIndex(Index(['Beam_group1'], dtype='object', name='beam_group'))
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - ping_timePandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:30.276106', '2023-08-11 16:57:33.288034', '2023-08-11 16:57:36.293041', '2023-08-11 16:57:39.277018', '2023-08-11 16:57:42.277037', '2023-08-11 16:57:44.287659', '2023-08-11 16:57:46.277308', '2023-08-11 16:57:48.287941', '2023-08-11 16:57:50.278605', ... '2023-08-11 17:05:11.287060', '2023-08-11 17:05:13.278721', '2023-08-11 17:05:15.277386', '2023-08-11 17:05:17.278019', '2023-08-11 17:05:19.278667', '2023-08-11 17:05:21.289291', '2023-08-11 17:05:23.292936', '2023-08-11 17:05:25.278637', '2023-08-11 17:05:27.281294', '2023-08-11 17:05:29.279934'], dtype='datetime64[ns]', name='ping_time', length=213, freq=None)) - range_samplePandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, ... 36188, 36189, 36190, 36191, 36192, 36193, 36194, 36195, 36196, 36197], dtype='int64', name='range_sample', length=36198)) - transmit_samplePandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, ... 138, 139, 140, 141, 142, 143, 144, 145, 146, 147], dtype='int64', name='transmit_sample', length=148))
- beam_mode :
- vertical
- conversion_equation_t :
- type_3
<xarray.DatasetView> Size: 465MB Dimensions: (channel: 5, ping_time: 213, range_sample: 36198, transmit_sample: 148, channel_all: 5, beam_group: 1) Coordinates: * channel_all (channel_all) <U25 500B 'WBT 400140-15 ES1... * beam_group (beam_group) <U11 44B 'Beam_group1' * channel (channel) <U25 500B 'WBT 400140-15 ES120-7... * ping_time (ping_time) datetime64[ns] 2kB 2023-08-11T... * range_sample (range_sample) int64 290kB 0 1 ... 36197 * transmit_sample (transmit_sample) int64 1kB 0 1 2 ... 146 147 Data variables: (12/29) frequency_nominal (channel) float64 40B 1.2e+05 ... 2e+05 beam_type (channel) int64 40B 1 1 1 1 1 beamwidth_twoway_alongship (channel) float64 40B 6.63 10.92 ... 6.79 beamwidth_twoway_athwartship (channel) float64 40B 6.74 10.6 ... 6.81 6.07 beam_direction_x (channel) float64 40B nan nan nan nan nan beam_direction_y (channel) float64 40B nan nan nan nan nan ... ... transmit_power (channel, ping_time) float64 9kB 250.0 ...... transmit_duration_nominal (channel, ping_time) float32 4kB 0.001024 ... slope (channel, ping_time) float64 9kB 0.01628 .... channel_mode (channel, ping_time) int8 1kB 0 0 0 ... 0 0 0 transmit_type (channel, ping_time) <U3 13kB 'CW' ... 'CW' sample_time_offset (channel, ping_time) float64 9kB 0.0 ... 0.0 Attributes: beam_mode: vertical conversion_equation_t: type_3xarray.DatasetView -
- channel: 5
- pulse_length_bin: 5
- cal_channel_id: 3
- cal_frequency: 269
- WBT_filter_n: 47
- PC_filter_n: 123
- channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - pulse_length_bin(pulse_length_bin)int640 1 2 3 4
array([0, 1, 2, 3, 4])
- cal_frequency(cal_frequency)int6445000 45473 45947 ... 240478 241459
- long_name :
- Frequency of calibration parameter
- units :
- Hz
array([ 45000, 45473, 45947, ..., 239497, 240478, 241459])
- cal_channel_id(cal_channel_id)object'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- ID of channels containing broadband calibration information
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400145-15 ES200-7C_ES'], dtype=object)
- frequency_nominal(channel)float641.2e+05 1.8e+04 7e+04 3.8e+04 2e+05
- units :
- Hz
- long_name :
- Transducer frequency
- valid_min :
- 0.0
- standard_name :
- sound_frequency
array([120000., 18000., 70000., 38000., 200000.])
- sa_correction(channel, pulse_length_bin)float640.0 0.0 0.0 0.0 ... 0.0 0.0 -0.09
array([[ 0. , 0. , 0. , 0. , -0.12], [ 0. , -0.07, 0. , 0. , 0. ], [ 0. , 0. , 0. , -0.2 , 0. ], [ 0. , 0. , -0.07, 0. , 0. ], [ 0. , 0. , 0. , 0. , -0.09]]) - gain_correction(channel, pulse_length_bin)float6427.0 27.0 27.0 ... 26.0 26.0 27.55
array([[27. , 27. , 27. , 27. , 26.76], [20.3 , 23.08, 22.9 , 23. , 23. ], [27. , 27. , 27. , 28.1 , 27. ], [23. , 25. , 26.46, 25.5 , 25. ], [26. , 26. , 26. , 26. , 27.55]]) - pulse_length(channel, pulse_length_bin)float646.4e-05 0.000128 ... 0.001024
array([[6.400e-05, 1.280e-04, 2.560e-04, 5.120e-04, 1.024e-03], [5.120e-04, 1.024e-03, 2.048e-03, 4.096e-03, 8.192e-03], [1.280e-04, 2.560e-04, 5.120e-04, 1.024e-03, 2.048e-03], [2.560e-04, 5.120e-04, 1.024e-03, 2.048e-03, 4.096e-03], [6.400e-05, 1.280e-04, 2.560e-04, 5.120e-04, 1.024e-03]]) - impedance_transceiver(channel)int645400 5400 5400 5400 5400
- units :
- ohm
- long_name :
- Transceiver impedance
array([5400, 5400, 5400, 5400, 5400])
- receiver_sampling_frequency(channel)float641.5e+06 1.5e+06 ... 1.5e+06 1.5e+06
- units :
- Hz
- long_name :
- Receiver sampling frequency
array([1500000., 1500000., 1500000., 1500000., 1500000.])
- transceiver_type(channel)<U3'WBT' 'WBT' 'WBT' 'WBT' 'WBT'
- long_name :
- Transceiver type
array(['WBT', 'WBT', 'WBT', 'WBT', 'WBT'], dtype='<U3')
- gain(cal_channel_id, cal_frequency)float64nan nan nan ... 28.41 28.33 28.35
array([[ nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 25.45, 25.44, 25.4 , 25.63, 25.86, 25.85, 25.85, 26.07, 26.16, 26.01, 25.9 , 25.92, 25.86, 25.77, 25.8 , 25.89, 25.99, 26.03, 26.11, 26.23, 26.24, 26.17, 26.07, 26.02, 25.93, 25.86, 25.87, 25.92, 26.07, 26.17, 26.29, 26.35, 26.38, 26.38, 26.26, 26.21, 26.16, 26.23, 26.25, 26.45, 26.64, 26.83, 26.97, 27.11, 27.24, 27.25, 27.27, 27.13, 27.05, 26.94, 26.88, 26.87, 26.92, 27.13, 27.29, 27.47, 27.62, 27.66, 27.66, 27.57, 27.54, 27.45, 27.37, 27.39, 27.52, 27.67, 27.87, 28.01, 28.12, 28.08, 28. , 27.86, 27.52, 26.98, 26.93, 26.93, 27.01, 27.16, 27.39, 27.67, 27.97, 28.13, 28.09, 27.98, 27.86, 27.77, 27.75, 27.71, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 24.16, 24.84, 25.38, 28.08, 24.69, 25.39, 25.48, 25.8 , 26.26, 26.42, 26.71, 27.17, 27.17, 26.95, 26.61, 26.83, 27.31, 27.62, 27.57, 27.29, 27.07, 27.44, 27.7 , 27.83, 27.66, 27.31, 27.31, 27.66, 27.98, 27.99, 27.73, 27.53, 27.72, 28.02, 28.29, 28.13, 27.84, 28.94, 28.81, 28.46, 28.19, 28.44, 28.89, 29.07, 28.78, 27.5 , 26.81, 27.19, 27.7 , 28.74, 28.36, 27.99, 28.06, 28.44, 28.68, 28.53, 27.93, 27.9 , 28.31, 28.77, 28.81, 28.41, 28.33, 28.35]]) - impedance_transducer(cal_channel_id, cal_frequency)float64nan nan nan nan ... 75.0 75.0 75.0
array([[nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75., 75.]]) - phase(cal_channel_id, cal_frequency)float64nan nan nan nan ... 0.0 0.0 0.0 0.0
array([[nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.]]) - beamwidth_alongship(cal_channel_id, cal_frequency)float64nan nan nan nan ... 6.25 5.95 5.85
array([[ nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 7.81, 7.99, 8.72, 8.31, 7.67, 8. , 8.64, 8.02, 7.65, 8. , 8.23, 7.75, 7.43, 7.81, 7.83, 7.59, 7.59, 7.68, 7.76, 7.51, 7.46, 7.52, 7.45, 7.33, 7.34, 7.42, 7.41, 7.46, 7.53, 7.37, 7.37, 7.47, 7.38, 7.18, 7.26, 7.36, 7.17, 7.12, 7.1 , 7.02, 6.96, 6.67, 6.54, 6.49, 6.41, 6.31, 6.3 , 6.34, 6.34, 6.33, 6.44, 6.52, 6.42, 6.29, 6.31, 6.28, 6.12, 6.06, 6.1 , 6.14, 6.04, 6.09, 6.21, 6.17, 6.01, 6.03, 6.04, 5.86, 5.8 , 5.84, 5.83, 5.88, 5.83, 5.5 , 5.45, 5.52, 5.45, 5.36, 5.3 , 5.29, 5.31, 5.24, 5.25, 5.25, 5.19, 5.2 , 5.16, 5.13, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 11.32, 9.04, 10.95, 8.84, 8.95, 8.15, 8.08, 8.66, 8.2 , 8.82, 8.73, 8.15, 8.46, 8.12, 8.79, 8.15, 8.15, 7.74, 7.91, 7.59, 8.13, 7.38, 7.62, 7.38, 7.35, 7.03, 7.15, 7.02, 6.87, 7.23, 7.28, 7.05, 7.03, 6.77, 7.39, 7.32, 7.03, 6.61, 6.67, 6.51, 6.81, 6.46, 6.3 , 6.24, 6.7 , 6.33, 6.37, 6.02, 7.76, 6.17, 6.35, 6.1 , 6.22, 6.06, 6.15, 6.24, 5.78, 6.6 , 6.15, 5.98, 6.6 , 6.25, 5.95, 5.85]]) - beamwidth_athwartship(cal_channel_id, cal_frequency)float64nan nan nan nan ... 5.91 5.55 5.38
array([[ nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 8.17, 8.17, 8.39, 8.06, 7.96, 8.09, 8.03, 8.03, 7.88, 7.8 , 7.81, 7.77, 7.6 , 7.63, 7.69, 7.66, 7.59, 7.6 , 7.62, 7.46, 7.43, 7.4 , 7.36, 7.32, 7.28, 7.3 , 7.31, 7.3 , 7.27, 7.29, 7.28, 7.13, 7.08, 7.16, 7.11, 7.16, 7.06, 7.05, 7.03, 7. , 6.9 , 6.63, 6.5 , 6.49, 6.42, 6.34, 6.34, 6.27, 6.27, 6.25, 6.23, 6.23, 6.27, 6.22, 6.17, 6.14, 6.08, 5.99, 6. , 5.96, 5.85, 5.87, 5.92, 5.95, 6. , 5.94, 5.87, 5.79, 5.83, 5.78, 5.68, 5.66, 5.72, 5.39, 5.42, 5.36, 5.32, 5.33, 5.28, 5.26, 5.18, 5.19, 5.19, 5.17, 5.15, 5.07, 5. , 5.07, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 7.77, 7.06, 7.62, 7.8 , 7.79, 7.63, 7.1 , 7.52, 7.14, 7.74, 8.09, 7.69, 7.88, 7.45, 7.82, 7.15, 7.16, 7.2 , 7.33, 7.21, 7.25, 6.6 , 7.05, 6.88, 6.75, 6.89, 6.5 , 6.58, 6.45, 6.43, 6.57, 6.32, 6.31, 6.44, 6.28, 6.73, 6.35, 6.01, 6.17, 6.11, 5.8 , 5.9 , 5.74, 5.88, 6.13, 6.05, 6.02, 5.78, 5.68, 5.83, 5.95, 5.69, 5.86, 5.8 , 5.67, 5.71, 6.11, 5.77, 5.55, 5.49, 5.29, 5.91, 5.55, 5.38]]) - angle_offset_alongship(cal_channel_id, cal_frequency)float64nan nan nan ... -0.12 -0.07 0.02
array([[ nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 0.02, 0.07, -0.11, -0.17, -0.03, 0. , -0.17, -0.17, -0.02, 0.05, -0.12, -0.09, 0.07, -0.05, -0.16, -0.12, -0.11, -0.12, -0.15, -0.12, -0.09, -0.11, -0.09, -0.08, -0.09, -0.11, -0.14, -0.15, -0.18, -0.19, -0.15, -0.15, -0.14, -0.13, -0.12, -0.15, -0.12, -0.12, -0.14, -0.13, -0.13, -0.17, -0.16, -0.15, -0.14, -0.13, -0.12, -0.12, -0.12, -0.14, -0.17, -0.18, -0.21, -0.22, -0.21, -0.18, -0.18, -0.16, -0.13, -0.15, -0.13, -0.15, -0.17, -0.19, -0.2 , -0.21, -0.22, -0.21, -0.21, -0.2 , -0.15, -0.17, -0.16, -0.18, -0.17, -0.2 , -0.19, -0.21, -0.21, -0.19, -0.2 , -0.19, -0.16, -0.14, -0.14, -0.18, -0.18, -0.16, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 0.9 , 0.17, 0.22, -0.04, -0.11, -0.22, -0.17, -0.1 , -0.07, -0.03, 0.05, -0.02, -0.07, -0.06, -0.03, 0.03, 0.03, -0.02, -0.08, -0.1 , 0.01, 0.01, -0.01, -0.15, -0.12, -0.2 , -0.05, -0.12, -0.1 , -0.16, -0.22, -0.2 , -0.07, 0.07, -0.09, -0.13, -0.17, 0.05, -0.02, -0.07, -0.13, 0.08, 0.07, -0.02, 0.03, -0.12, 0.04, 0.02, 0.17, -0.04, -0.13, -0.02, 0.12, 0.04, -0.01, -0.07, -0.02, -0.09, 0. , -0.02, -0.25, -0.12, -0.07, 0.02]]) - angle_offset_athwartship(cal_channel_id, cal_frequency)float64nan nan nan nan ... 0.11 0.08 0.09
array([[ nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, -0.12, -0.1 , -0.07, -0.04, 0.02, 0. , 0.01, 0.08, 0.04, -0.03, 0.01, 0.06, 0. , 0.04, 0.09, 0.08, 0.1 , 0.14, 0.12, 0.1 , 0.08, 0.07, 0.05, 0.06, 0.04, 0.06, 0.12, 0.12, 0.12, 0.15, 0.13, 0.09, 0.07, 0.07, 0.05, 0.07, 0.08, 0.1 , 0.14, 0.18, 0.17, 0.15, 0.16, 0.19, 0.18, 0.17, 0.16, 0.13, 0.12, 0.12, 0.13, 0.15, 0.16, 0.16, 0.17, 0.19, 0.2 , 0.17, 0.14, 0.12, 0.12, 0.12, 0.17, 0.19, 0.21, 0.19, 0.16, 0.17, 0.18, 0.16, 0.13, 0.11, 0.11, 0.15, 0.16, 0.18, 0.19, 0.19, 0.18, 0.2 , 0.18, 0.16, 0.17, 0.16, 0.15, 0.15, 0.15, 0.17, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, 0.18, 0.1 , 0.14, 0.08, 0.2 , 0.14, 0.11, 0.06, 0.14, 0.1 , 0.17, 0.12, 0.09, 0.07, 0.05, 0.14, 0.14, 0.13, 0.09, 0.08, 0.09, 0.14, 0.16, 0.11, 0.08, 0.03, 0.09, 0.12, 0.11, 0.08, 0.06, 0.06, 0.08, 0.15, 0.1 , 0.09, 0.04, 0.17, 0.14, 0.11, 0.17, 0.2 , 0.19, 0.14, 0.11, 0.16, 0.13, 0.12, 0.13, 0.18, 0.11, 0.08, 0.06, 0.14, 0.14, 0.13, 0.13, 0.09, 0.13, 0.09, 0.03, 0.11, 0.08, 0.09]]) - WBT_filter_i(channel, WBT_filter_n)float320.0004271 0.002565 ... nan nan
- long_name :
- Wideband transceiver filter coefficients (imaginary part)
array([[ 4.27060440e-04, 2.56522908e-03, 7.59602245e-03, 1.35238143e-02, 1.24657992e-02, -7.21828081e-03, -5.18517978e-02, -1.11618504e-01, -1.58596575e-01, -1.60175383e-01, -1.01497099e-01, 0.00000000e+00, 1.01497099e-01, 1.60175383e-01, 1.58596575e-01, 1.11618504e-01, 5.18517978e-02, 7.21828081e-03, -1.24657992e-02, -1.35238143e-02, -7.59602245e-03, -2.56522908e-03, -4.27060440e-04, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan], [-1.80988136e-04, -4.21589793e-04, -8.79614206e-04, -1.61740114e-03, -2.71854317e-03, -4.26115468e-03, -6.30566804e-03, -8.88137147e-03, -1.19733466e-02, -1.55117726e-02, -1.93656068e-02, -2.33423840e-02, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan], [-1.29707449e-04, -2.33587972e-03, -7.35451514e-03, -7.13891769e-03, 1.37733202e-02, 5.47739267e-02, 7.63969049e-02, 2.56477650e-02, -9.34050456e-02, -1.88668311e-01, -1.56567663e-01, 0.00000000e+00, 1.56567663e-01, 1.88668311e-01, 9.34050456e-02, -2.56477650e-02, -7.63969049e-02, -5.47739267e-02, -1.37733202e-02, 7.13891769e-03, 7.35451514e-03, 2.33587972e-03, 1.29707449e-04, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan]], dtype=float32) - WBT_filter_r(channel, WBT_filter_n)float320.0004548 0.0008335 ... nan nan
- long_name :
- Wideband transceiver filter coefficients (real part)
array([[ 4.54773195e-04, 8.33493425e-04, -1.44901872e-03, -1.11878719e-02, -3.14850174e-02, -5.71385771e-02, -7.13678747e-02, -5.25236800e-02, 9.97805130e-03, 1.01650387e-01, 1.84622541e-01, 2.18127519e-01, 1.84622541e-01, 1.01650387e-01, 9.97805130e-03, -5.25236800e-02, -7.13678747e-02, -5.71385771e-02, -3.14850174e-02, -1.11878719e-02, -1.44901872e-03, 8.33493425e-04, 4.54773195e-04, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan], [-2.98325904e-05, -3.71809219e-05, -1.10541396e-05, 1.01758254e-04, 3.78196331e-04, 9.24408610e-04, 1.87500881e-03, 3.38791404e-03, 5.63422916e-03, 8.78328830e-03, 1.29837319e-02, 1.83422714e-02, ... nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan], [-6.10225601e-04, -1.34862075e-03, 2.38962681e-03, 1.60342716e-02, 3.09353843e-02, 1.77971274e-02, -4.41077724e-02, -1.20663248e-01, -1.28561020e-01, -1.98298376e-02, 1.40974149e-01, 2.18127519e-01, 1.40974149e-01, -1.98298376e-02, -1.28561020e-01, -1.20663248e-01, -4.41077724e-02, 1.77971274e-02, 3.09353843e-02, 1.60342716e-02, 2.38962681e-03, -1.34862075e-03, -6.10225601e-04, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan]], dtype=float32) - WBT_decimation(channel)int646 6 6 6 6
- long_name :
- Wideband transceiver decimation factor
array([6, 6, 6, 6, 6])
- PC_filter_i(channel, PC_filter_n)float32-2.988e-05 2.629e-05 ... -2.893e-05
- long_name :
- Pulse compression filter coefficients (imaginary part)
array([[-2.98759260e-05, 2.62850190e-05, -3.64410589e-05, 4.77569047e-05, -5.94764861e-05, 7.04559498e-05, -7.91147031e-05, 8.34040766e-05, -8.08000113e-05, 6.83263133e-05, -4.26138067e-05, 2.45083441e-18, 6.33284653e-05, -1.51150060e-04, 2.66981806e-04, -4.13817208e-04, 5.93837001e-04, -8.08104523e-04, 1.05625938e-03, -1.33622682e-03, 1.64396141e-03, -1.97324320e-03, 2.31554708e-03, -2.66000303e-03, 2.99346284e-03, -3.30068497e-03, 3.56464717e-03, -3.76698514e-03, 3.88855790e-03, -3.91012197e-03, 3.81310424e-03, -3.58043844e-03, 3.19744600e-03, -2.65271310e-03, 1.93893374e-03, -1.05367170e-03, 2.65581291e-17, 1.21301925e-03, -2.57005356e-03, 4.04944457e-03, -5.62339835e-03, 7.25843804e-03, -8.91611166e-03, 1.05539467e-02, -1.21266339e-02, 1.35873929e-02, -1.48894852e-02, 1.59878153e-02, -1.68405622e-02, 1.74107682e-02, -1.76678319e-02, 1.75888222e-02, -1.71595793e-02, 1.63755212e-02, -1.52421333e-02, 1.37751121e-02, -1.20001305e-02, 9.95224994e-03, -7.67497160e-03, 5.21897525e-03, ... -1.23351933e-02, 1.22546479e-02, 1.96472947e-02, -9.25248698e-17, -1.91380773e-02, -1.16274506e-02, 1.13999462e-02, 1.80363040e-02, -1.67636873e-16, -1.71061084e-02, -1.02540310e-02, 9.91820917e-03, 1.54795069e-02, -2.68487368e-16, -1.42816063e-02, -8.44205078e-03, 8.05107132e-03, 1.23873567e-02, -2.23871602e-16, -1.11014657e-02, -6.46575913e-03, 6.07439363e-03, 9.20466427e-03, -1.72641713e-16, -7.99549837e-03, -4.58272407e-03, 4.23559407e-03, 6.31225714e-03, -2.08926606e-16, -5.29763242e-03, -2.98283389e-03, 2.70704995e-03, 3.95944994e-03, -1.32215755e-16, -3.19574471e-03, -1.76299294e-03, 1.56660262e-03, 2.24192766e-03, -7.52219586e-17, -1.72795553e-03, -9.30224778e-04, 8.05766671e-04, 1.12271798e-03, -3.76065853e-17, -8.16939690e-04, -4.26290586e-04, 3.57247365e-04, 4.80550691e-04, -1.58999218e-17, -3.23363201e-04, -1.61491145e-04, 1.29013599e-04, 1.64652651e-04, -5.26718670e-18, -9.78860553e-05, -4.53715948e-05, 3.32463424e-05, 3.83029255e-05, -1.88707877e-18, -2.89260643e-05]], dtype=float32) - PC_filter_r(channel, PC_filter_n)float32-5.649e-06 8.591e-06 ... 9.147e-06
- long_name :
- Pulse compression filter coefficients (real part)
array([[-5.64898892e-06, 8.59066949e-06, -1.70977128e-05, 3.03576016e-05, -4.91530809e-05, 7.50781110e-05, -1.08841901e-04, 1.51761589e-04, -2.04027383e-04, 2.66163552e-04, -3.37272882e-04, 4.16491675e-04, -5.01246250e-04, 5.88740746e-04, -6.74268929e-04, 7.52780819e-04, -8.17296386e-04, 8.60594038e-04, -8.73763580e-04, 8.48045514e-04, -7.73539417e-04, 6.41195744e-04, -4.41664073e-04, 1.67403356e-04, 1.88382866e-04, -6.29589194e-04, 1.15827424e-03, -1.77255855e-03, 2.46780389e-03, -3.23468423e-03, 4.06059343e-03, -4.92800074e-03, 5.81618259e-03, -6.69993786e-03, 7.55169848e-03, -8.34061950e-03, 9.03505739e-03, -9.60198604e-03, 1.00097479e-02, -1.02276783e-02, 1.02289738e-02, -9.99033265e-03, 9.49474424e-03, -8.73093400e-03, 7.69584626e-03, -6.39369152e-03, 4.83793719e-03, -3.04978876e-03, 1.05956849e-03, 1.09544292e-03, -3.37026897e-03, 5.71500463e-03, -8.07463471e-03, 1.03922710e-02, -1.26093393e-02, 1.46690542e-02, -1.65167134e-02, 1.81031264e-02, -1.93847157e-02, 2.03266162e-02, ... -1.69781893e-02, -1.68673284e-02, 6.38354104e-03, 2.04155985e-02, 6.21808646e-03, -1.60040651e-02, -1.56909321e-02, 5.86009864e-03, 1.84937380e-02, 5.55786025e-03, -1.41137149e-02, -1.36514958e-02, 5.02934493e-03, 1.56554859e-02, 4.64012334e-03, -1.16197383e-02, -1.10815996e-02, 4.02464438e-03, 1.23485457e-02, 3.60683305e-03, -8.89960583e-03, -8.36093724e-03, 2.99052498e-03, 9.03475937e-03, 2.59764330e-03, -6.30783010e-03, -5.83004672e-03, 2.05072504e-03, 6.09115651e-03, 1.72105350e-03, -4.10577049e-03, -3.72618623e-03, 1.28625159e-03, 3.74784158e-03, 1.03810872e-03, -2.42680311e-03, -2.15649512e-03, 7.28194835e-04, 2.07453989e-03, 5.61195135e-04, -1.28059613e-03, -1.10929436e-03, 3.64541571e-04, 1.01004471e-03, 2.65188166e-04, -5.86990267e-04, -4.91960440e-04, 1.55888760e-04, 4.16190072e-04, 1.04815445e-04, -2.22525108e-04, -1.77823618e-04, 5.32472586e-05, 1.34346395e-04, 3.15534780e-05, -6.27002737e-05, -4.60112933e-05, 1.21937455e-05, 2.73860878e-05, 9.14701923e-06]], dtype=float32) - PC_decimation(channel)int6410 7 12 10 8
- long_name :
- Pulse compression decimation factor
array([10, 7, 12, 10, 8])
- config_xml()<U74340'<?xml version="1.0" encoding="u...
array('<?xml version="1.0" encoding="utf-8" ?>\n<Configuration>\n <Header Copyright="Copyright(c) Kongsberg Maritime AS, Norway" ApplicationName="EK80" Version="21.15.2.0" FileFormatVersion="1.27" TimeBias="0" />\n <ActivePingMode Mode="Direct" />\n <Transceivers MergeOperation="AddNodeTree">\n <Transceiver TransceiverName="WBT 400141" IPAddress="157.237.15.100" MarketSegment="Scientific" SerialNumber="400141" Impedance="5400" Multiplexing="0" RxSampleFrequency="1500000" TransceiverType="WBT" EthernetAddress="009072061b0d" Version="[0] Ethernet: 00:90:72:06:1B:0D
[1] Parts-list: WBT 371790/F
[2] Product: WBT
IP Address: 157.237.15.100
Subnet mask: 255.255.0.0
Default gateway: 157.237.15.1
Serial number: 400141
Embedded software: Rev. 2.54
FPGA TX firmware: Rev. 5
FPGA RX firmware: Rev. 7
CH1: 509W CH2: 498W CH3: 494W CH4: 499W
TRD1: Unable to detect transducer
TRD2: Unable to detect transducer
TRD3: Unable to detect transducer
TRD4: Unable to detect transducer
" TransceiverSoftwareVersion="2.54" TransceiverNumber="1">\n <Channels>\n <Channel ChannelID="WBT 400141-15 ES18_ES" LogicalChannelID="WBT 400141-15 ES18" ChannelIdShort="ES18 Shimada" MaxTxPowerTransceiver="2000" HWChannelConfiguration="15" PulseDuration="0.000512;0.001024;0.002048;0.004096;0.008192">\n <Transducer TransducerName="ES18" ArticleNumber="KSV-088694" SerialNumber="0" Frequency="18000" FrequencyMinimum="18000" FrequencyMaximum="18000" MaxTxPowerTransducer="1000" Gain="20.3;23.08;22.9;23;23" SaCorrection="0;-0.07;0;0;0" EquivalentBeamAngle="-17" DirectivityDropAt2XBeamWidth="0" AngleSensitivityAlongship="15.5" AngleSensitivityAthwartship="15.5" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="-0.05" BeamWidthAlongship="10.92" BeamWidthAthwartship="10.6" BeamType="1" />\n </Channel>\n </Channels>\n </Transceiver>\n <Transceiver TransceiverName="WBT 400143" IPAddress="157.237.15.104" MarketSegment="Scientific" SerialNumber="400143" Impedance="5400" Multiplexing="0" RxSampleFrequency="1500000" TransceiverType="WBT" EthernetAddress="009072061b0f" Version="[0] Ethernet: 00:90:72:06:1B:0F
[1] Parts-list: WBT 371790/F
[2] Product: WBT
IP Address: 157.237.15.104
Subnet mask: 255.255.0.0
Default gateway: 157.237.15.1
Serial number: 400143
Embedded software: Rev. 2.54
FPGA TX firmware: Rev. 5
FPGA RX firmware: Rev. 7
CH1: 514W CH2: 499W CH3: 491W CH4: 502W
TRD1: Unable to detect transducer
TRD2: Unable to detect transducer
TRD3: Unable to detect transducer
TRD4: Unable to detect transducer
" TransceiverSoftwareVersion="2.54" TransceiverNumber="2">\n <Channels>\n <Channel ChannelID="WBT 400143-15 ES38B_ES" LogicalChannelID="WBT 400143-15 ES38B" ChannelIdShort="ES 38 Shimada" MaxTxPowerTransceiver="2000" HWChannelConfiguration="15" PulseDuration="0.000256;0.000512;0.001024;0.002048;0.004096">\n <Transducer TransducerName="ES38B" SerialNumber="0" Frequency="38000" FrequencyMinimum="38000" FrequencyMaximum="38000" MaxTxPowerTransducer="2000" Gain="23;25;26.46;25.5;25" SaCorrection="0;0;-0.07;0;0" EquivalentBeamAngle="-20.7" DirectivityDropAt2XBeamWidth="0" AngleSensitivityAlongship="23" AngleSensitivityAthwartship="23" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="-0.17" BeamWidthAlongship="6.71" BeamWidthAthwartship="6.81" BeamType="1" />\n </Channel>\n </Channels>\n </Transceiver>\n <Transceiver TransceiverName="WBT 400142" IPAddress="157.237.15.103" MarketSegment="Scientific" SerialNumber="400142" Impedance="5400" Multiplexing="0" RxSampleFrequency="1500000" TransceiverType="WBT" EthernetAddress="009072061b0e" Version="[0] Ethernet: 00:90:72:06:1B:0E
[1] Parts-list: WBT 371790/F
[2] Product: WBT
IP Address: 157.237.15.103
Subnet mask: 255.255.0.0
Default gateway: 157.237.15.1
Serial number: 400142
Embedded software: Rev. 2.54
FPGA TX firmware: Rev. 5
FPGA RX firmware: Rev. 7
CH1: 512W CH2: 502W CH3: 490W CH4: 509W
TRD1: Unable to detect transducer
TRD2: Unable to detect transducer
TRD3: Unable to detect transducer
TRD4: Unable to detect transducer
" TransceiverSoftwareVersion="2.54" TransceiverNumber="3">\n <Channels>\n <Channel ChannelID="WBT 400142-15 ES70-7C_ES" LogicalChannelID="WBT 400142-15 ES70-7C" ChannelIdShort="ES70 Shimada" MaxTxPowerTransceiver="2000" HWChannelConfiguration="15" PulseDuration="0.000128;0.000256;0.000512;0.001024;0.002048" PulseDurationFM="0.000512;0.001024;0.002048;0.004096;0.008192">\n <Transducer TransducerName="ES70-7C" ArticleNumber="KSV-203678" SerialNumber="0" Frequency="70000" FrequencyMinimum="45000" FrequencyMaximum="90000" MaxTxPowerTransducer="750" Gain="27;27;27;28.1;27" SaCorrection="0;0;0;-0.2;0" EquivalentBeamAngle="-20.7" DirectivityDropAt2XBeamWidth="0" AngleSensitivityAlongship="23" AngleSensitivityAthwartship="23" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0" BeamWidthAlongship="6.55" BeamWidthAthwartship="6.61" BeamType="1">\n <FrequencyPar Frequency="45000" Gain="22.04" Impedance="75" Phase="0" BeamWidthAlongship="10.13" BeamWidthAthwartship="9.87" AngleOffsetAlongship="0.41" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="45473" Gain="22.46" Impedance="75" Phase="0" BeamWidthAlongship="10.23" BeamWidthAthwartship="10.09" AngleOffsetAlongship="0.48" AngleOffsetAthwartship="-0.01" />\n <FrequencyPar Frequency="45947" Gain="22.96" Impedance="75" Phase="0" BeamWidthAlongship="10.25" BeamWidthAthwartship="10.07" AngleOffsetAlongship="0.5" AngleOffsetAthwartship="-0.14" />\n <FrequencyPar Frequency="46420" Gain="23.46" Impedance="75" Phase="0" BeamWidthAlongship="9.87" BeamWidthAthwartship="9.9" AngleOffsetAlongship="0.48" AngleOffsetAthwartship="-0.12" />\n <FrequencyPar Frequency="46894" Gain="23.87" Impedance="75" Phase="0" BeamWidthAlongship="9.72" BeamWidthAthwartship="9.8" AngleOffsetAlongship="0.49" AngleOffsetAthwartship="-0.13" />\n <FrequencyPar Frequency="47367" Gain="24.15" Impedance="75" Phase="0" BeamWidthAlongship="9.66" BeamWidthAthwartship="9.64" AngleOffsetAlongship="0.42" AngleOffsetAthwartship="-0.1" />\n <FrequencyPar Frequency="47841" Gain="24.36" Impedance="75" Phase="0" BeamWidthAlongship="9.51" BeamWidthAthwartship="9.53" AngleOffsetAlongship="0.37" AngleOffsetAthwartship="-0.11" />\n <FrequencyPar Frequency="48314" Gain="24.45" Impedance="75" Phase="0" BeamWidthAlongship="9.39" BeamWidthAthwartship="9.39" AngleOffsetAlongship="0.32" AngleOffsetAthwartship="-0.12" />\n <FrequencyPar Frequency="48788" Gain="24.48" Impedance="75" Phase="0" BeamWidthAlongship="9.46" BeamWidthAthwartship="9.19" AngleOffsetAlongship="0.27" AngleOffsetAthwartship="-0.08" />\n <FrequencyPar Frequency="49261" Gain="24.42" Impedance="75" Phase="0" BeamWidthAlongship="9.72" BeamWidthAthwartship="9.09" AngleOffsetAlongship="0.25" AngleOffsetAthwartship="-0.08" />\n <FrequencyPar Frequency="49735" Gain="24.41" Impedance="75" Phase="0" BeamWidthAlongship="9.31" BeamWidthAthwartship="9.11" AngleOffsetAlongship="0.13" AngleOffsetAthwartship="-0.06" />\n <FrequencyPar Frequency="50208" Gain="24.38" Impedance="75" Phase="0" BeamWidthAlongship="9.23" BeamWidthAthwartship="8.97" AngleOffsetAlongship="0.1" AngleOffsetAthwartship="-0.05" />\n <FrequencyPar Frequency="50682" Gain="24.38" Impedance="75" Phase="0" BeamWidthAlongship="9.13" BeamWidthAthwartship="8.88" AngleOffsetAlongship="0.08" AngleOffsetAthwartship="-0.01" />\n <FrequencyPar Frequency="51155" Gain="24.43" Impedance="75" Phase="0" BeamWidthAlongship="8.82" BeamWidthAthwartship="8.85" AngleOffsetAlongship="-0.01" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="51629" Gain="24.48" Impedance="75" Phase="0" BeamWidthAlongship="8.95" BeamWidthAthwartship="8.8" AngleOffsetAlongship="-0.01" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="52102" Gain="24.62" Impedance="75" Phase="0" BeamWidthAlongship="8.7" BeamWidthAthwartship="8.64" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="52576" Gain="24.7" Impedance="75" Phase="0" BeamWidthAlongship="8.54" BeamWidthAthwartship="8.48" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="53049" Gain="24.78" Impedance="75" Phase="0" BeamWidthAlongship="8.57" BeamWidthAthwartship="8.41" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="53523" Gain="24.8" Impedance="75" Phase="0" BeamWidthAlongship="8.67" BeamWidthAthwartship="8.37" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="53996" Gain="24.86" Impedance="75" Phase="0" BeamWidthAlongship="8.49" BeamWidthAthwartship="8.29" AngleOffsetAlongship="-0.04" AngleOffsetAthwartship="0" />\n <FrequencyPar Frequency="54469" Gain="24.88" Impedance="75" Phase="0" BeamWidthAlongship="8.24" BeamWidthAthwartship="8.28" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="-0.02" />\n <FrequencyPar Frequency="54943" Gain="24.94" Impedance="75" Phase="0" BeamWidthAlongship="8.35" BeamWidthAthwartship="8.22" AngleOffsetAlongship="0" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="55416" Gain="25.01" Impedance="75" Phase="0" BeamWidthAlongship="8.26" BeamWidthAthwartship="8.16" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="55890" Gain="25.08" Impedance="75" Phase="0" BeamWidthAlongship="8.21" BeamWidthAthwartship="8.15" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0" />\n <FrequencyPar Frequency="56363" Gain="25.23" Impedance="75" Phase="0" BeamWidthAlongship="8.11" BeamWidthAthwartship="8.01" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0" />\n <FrequencyPar Frequency="56837" Gain="25.37" Impedance="75" Phase="0" BeamWidthAlongship="8.06" BeamWidthAthwartship="7.84" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="57310" Gain="25.54" Impedance="75" Phase="0" BeamWidthAlongship="7.93" BeamWidthAthwartship="7.79" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="57784" Gain="25.66" Impedance="75" Phase="0" BeamWidthAlongship="7.9" BeamWidthAthwartship="7.76" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="58257" Gain="25.79" Impedance="75" Phase="0" BeamWidthAlongship="7.9" BeamWidthAthwartship="7.71" AngleOffsetAlongship="-0.04" AngleOffsetAthwartship="-0.01" />\n <FrequencyPar Frequency="58731" Gain="25.92" Impedance="75" Phase="0" BeamWidthAlongship="7.62" BeamWidthAthwartship="7.59" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="59204" Gain="25.99" Impedance="75" Phase="0" BeamWidthAlongship="7.75" BeamWidthAthwartship="7.52" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="59678" Gain="26.05" Impedance="75" Phase="0" BeamWidthAlongship="7.73" BeamWidthAthwartship="7.52" AngleOffsetAlongship="-0.06" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="60151" Gain="26.14" Impedance="75" Phase="0" BeamWidthAlongship="7.36" BeamWidthAthwartship="7.5" AngleOffsetAlongship="-0.04" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="60625" Gain="26.23" Impedance="75" Phase="0" BeamWidthAlongship="7.45" BeamWidthAthwartship="7.39" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="61098" Gain="26.3" Impedance="75" Phase="0" BeamWidthAlongship="7.45" BeamWidthAthwartship="7.31" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="61572" Gain="26.46" Impedance="75" Phase="0" BeamWidthAlongship="7.28" BeamWidthAthwartship="7.17" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="62045" Gain="26.56" Impedance="75" Phase="0" BeamWidthAlongship="7.18" BeamWidthAthwartship="7.14" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="62519" Gain="26.7" Impedance="75" Phase="0" BeamWidthAlongship="7.18" BeamWidthAthwartship="7.07" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="62992" Gain="26.83" Impedance="75" Phase="0" BeamWidthAlongship="7.19" BeamWidthAthwartship="6.94" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="63465" Gain="26.96" Impedance="75" Phase="0" BeamWidthAlongship="7.17" BeamWidthAthwartship="6.87" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="63939" Gain="27.02" Impedance="75" Phase="0" BeamWidthAlongship="7.07" BeamWidthAthwartship="6.96" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="64412" Gain="27.09" Impedance="75" Phase="0" BeamWidthAlongship="7.05" BeamWidthAthwartship="6.96" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0" />\n <FrequencyPar Frequency="64886" Gain="27.15" Impedance="75" Phase="0" BeamWidthAlongship="6.96" BeamWidthAthwartship="6.85" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="65359" Gain="27.22" Impedance="75" Phase="0" BeamWidthAlongship="6.79" BeamWidthAthwartship="6.82" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="65833" Gain="27.25" Impedance="75" Phase="0" BeamWidthAlongship="6.9" BeamWidthAthwartship="6.8" AngleOffsetAlongship="-0.06" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="66306" Gain="27.32" Impedance="75" Phase="0" BeamWidthAlongship="6.78" BeamWidthAthwartship="6.79" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="66780" Gain="27.38" Impedance="75" Phase="0" BeamWidthAlongship="6.7" BeamWidthAthwartship="6.73" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="67253" Gain="27.46" Impedance="75" Phase="0" BeamWidthAlongship="6.72" BeamWidthAthwartship="6.6" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="67727" Gain="27.58" Impedance="75" Phase="0" BeamWidthAlongship="6.7" BeamWidthAthwartship="6.53" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="68200" Gain="27.69" Impedance="75" Phase="0" BeamWidthAlongship="6.57" BeamWidthAthwartship="6.5" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="68674" Gain="27.79" Impedance="75" Phase="0" BeamWidthAlongship="6.59" BeamWidthAthwartship="6.47" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="69147" Gain="27.95" Impedance="75" Phase="0" BeamWidthAlongship="6.52" BeamWidthAthwartship="6.39" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="69621" Gain="28.09" Impedance="75" Phase="0" BeamWidthAlongship="6.3" BeamWidthAthwartship="6.28" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="70094" Gain="28.14" Impedance="75" Phase="0" BeamWidthAlongship="6.4" BeamWidthAthwartship="6.34" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="70568" Gain="28.16" Impedance="75" Phase="0" BeamWidthAlongship="6.38" BeamWidthAthwartship="6.32" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="71041" Gain="28.24" Impedance="75" Phase="0" BeamWidthAlongship="6.25" BeamWidthAthwartship="6.15" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="71515" Gain="28.3" Impedance="75" Phase="0" BeamWidthAlongship="6.16" BeamWidthAthwartship="6.05" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="71988" Gain="28.19" Impedance="75" Phase="0" BeamWidthAlongship="6.14" BeamWidthAthwartship="6.08" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="72461" Gain="28.21" Impedance="75" Phase="0" BeamWidthAlongship="6.14" BeamWidthAthwartship="6" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="72935" Gain="28.3" Impedance="75" Phase="0" BeamWidthAlongship="5.97" BeamWidthAthwartship="5.91" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="73408" Gain="28.26" Impedance="75" Phase="0" BeamWidthAlongship="6.06" BeamWidthAthwartship="5.87" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="73882" Gain="28.36" Impedance="75" Phase="0" BeamWidthAlongship="5.94" BeamWidthAthwartship="5.79" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="74355" Gain="28.49" Impedance="75" Phase="0" BeamWidthAlongship="5.83" BeamWidthAthwartship="5.79" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="74829" Gain="28.49" Impedance="75" Phase="0" BeamWidthAlongship="5.94" BeamWidthAthwartship="5.83" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="75302" Gain="28.52" Impedance="75" Phase="0" BeamWidthAlongship="5.91" BeamWidthAthwartship="5.77" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="75776" Gain="28.65" Impedance="75" Phase="0" BeamWidthAlongship="5.84" BeamWidthAthwartship="5.68" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="76249" Gain="28.7" Impedance="75" Phase="0" BeamWidthAlongship="5.7" BeamWidthAthwartship="5.66" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="76723" Gain="28.61" Impedance="75" Phase="0" BeamWidthAlongship="5.84" BeamWidthAthwartship="5.66" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="77196" Gain="28.59" Impedance="75" Phase="0" BeamWidthAlongship="5.74" BeamWidthAthwartship="5.63" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="77670" Gain="28.63" Impedance="75" Phase="0" BeamWidthAlongship="5.63" BeamWidthAthwartship="5.54" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="78143" Gain="28.64" Impedance="75" Phase="0" BeamWidthAlongship="5.69" BeamWidthAthwartship="5.49" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="78617" Gain="28.61" Impedance="75" Phase="0" BeamWidthAlongship="5.6" BeamWidthAthwartship="5.53" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="79090" Gain="28.62" Impedance="75" Phase="0" BeamWidthAlongship="5.6" BeamWidthAthwartship="5.45" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="79564" Gain="28.74" Impedance="75" Phase="0" BeamWidthAlongship="5.53" BeamWidthAthwartship="5.43" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="80037" Gain="28.89" Impedance="75" Phase="0" BeamWidthAlongship="5.49" BeamWidthAthwartship="5.45" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="80511" Gain="29.12" Impedance="75" Phase="0" BeamWidthAlongship="5.34" BeamWidthAthwartship="5.32" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="80984" Gain="29.24" Impedance="75" Phase="0" BeamWidthAlongship="5.35" BeamWidthAthwartship="5.27" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="81457" Gain="29.35" Impedance="75" Phase="0" BeamWidthAlongship="5.29" BeamWidthAthwartship="5.24" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="81931" Gain="29.4" Impedance="75" Phase="0" BeamWidthAlongship="5.21" BeamWidthAthwartship="5.16" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="82404" Gain="29.42" Impedance="75" Phase="0" BeamWidthAlongship="5.25" BeamWidthAthwartship="5.08" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="82878" Gain="29.42" Impedance="75" Phase="0" BeamWidthAlongship="5.18" BeamWidthAthwartship="5.12" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="83351" Gain="29.42" Impedance="75" Phase="0" BeamWidthAlongship="5.24" BeamWidthAthwartship="5.1" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="83825" Gain="29.51" Impedance="75" Phase="0" BeamWidthAlongship="5.12" BeamWidthAthwartship="5.04" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="84298" Gain="29.57" Impedance="75" Phase="0" BeamWidthAlongship="5.12" BeamWidthAthwartship="5" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="84772" Gain="29.68" Impedance="75" Phase="0" BeamWidthAlongship="5.07" BeamWidthAthwartship="4.92" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="85245" Gain="29.81" Impedance="75" Phase="0" BeamWidthAlongship="4.94" BeamWidthAthwartship="4.95" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="85719" Gain="29.9" Impedance="75" Phase="0" BeamWidthAlongship="5.03" BeamWidthAthwartship="4.93" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="86192" Gain="30.12" Impedance="75" Phase="0" BeamWidthAlongship="4.92" BeamWidthAthwartship="4.85" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="86666" Gain="30.35" Impedance="75" Phase="0" BeamWidthAlongship="4.85" BeamWidthAthwartship="4.79" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="87139" Gain="30.65" Impedance="75" Phase="0" BeamWidthAlongship="4.76" BeamWidthAthwartship="4.74" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="87613" Gain="31.01" Impedance="75" Phase="0" BeamWidthAlongship="4.68" BeamWidthAthwartship="4.67" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="88000" Gain="31.2" Impedance="75" Phase="0" BeamWidthAlongship="4.78" BeamWidthAthwartship="4.62" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.08" />\n </Transducer>\n </Channel>\n </Channels>\n </Transceiver>\n <Transceiver TransceiverName="WBT 400140" IPAddress="157.237.15.101" MarketSegment="Scientific" SerialNumber="400140" Impedance="5400" Multiplexing="0" RxSampleFrequency="1500000" TransceiverType="WBT" EthernetAddress="009072061b0c" Version="[0] Ethernet: 00:90:72:06:1B:0C
[1] Parts-list: WBT 371790/F
[2] Product: WBT
IP Address: 157.237.15.101
Subnet mask: 255.255.0.0
Default gateway: 157.237.15.1
Serial number: 400140
Embedded software: Rev. 2.54
FPGA TX firmware: Rev. 5
FPGA RX firmware: Rev. 7
CH1: 507W CH2: 489W CH3: 491W CH4: 486W
TRD1: Unable to detect transducer
TRD2: Unable to detect transducer
TRD3: Unable to detect transducer
TRD4: Unable to detect transducer
" TransceiverSoftwareVersion="2.54" TransceiverNumber="4">\n <Channels>\n <Channel ChannelID="WBT 400140-15 ES120-7C_ES" LogicalChannelID="WBT 400140-15 ES120-7C" ChannelIdShort="ES120 Shimada" MaxTxPowerTransceiver="2000" HWChannelConfiguration="15" PulseDuration="6.4E-05;0.000128;0.000256;0.000512;0.001024" PulseDurationFM="0.000512;0.001024;0.002048;0.004096;0.008192">\n <Transducer TransducerName="ES120-7C" ArticleNumber="KSV-204580" SerialNumber="0" Frequency="120000" FrequencyMinimum="90000" FrequencyMaximum="170000" MaxTxPowerTransducer="250" Gain="27;27;27;27;26.76" SaCorrection="0;0;0;0;-0.12" EquivalentBeamAngle="-20.7" DirectivityDropAt2XBeamWidth="0" AngleSensitivityAlongship="23" AngleSensitivityAthwartship="23" AngleOffsetAlongship="-0.04" AngleOffsetAthwartship="0.17" BeamWidthAlongship="6.63" BeamWidthAthwartship="6.74" BeamType="1">\n <FrequencyPar Frequency="95237" Gain="25.45" Impedance="75" Phase="0" BeamWidthAlongship="7.81" BeamWidthAthwartship="8.17" AngleOffsetAlongship="0.02" AngleOffsetAthwartship="-0.12" />\n <FrequencyPar Frequency="95699" Gain="25.44" Impedance="75" Phase="0" BeamWidthAlongship="7.99" BeamWidthAthwartship="8.17" AngleOffsetAlongship="0.07" AngleOffsetAthwartship="-0.1" />\n <FrequencyPar Frequency="96162" Gain="25.4" Impedance="75" Phase="0" BeamWidthAlongship="8.72" BeamWidthAthwartship="8.39" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="-0.07" />\n <FrequencyPar Frequency="96624" Gain="25.63" Impedance="75" Phase="0" BeamWidthAlongship="8.31" BeamWidthAthwartship="8.06" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="-0.04" />\n <FrequencyPar Frequency="97087" Gain="25.86" Impedance="75" Phase="0" BeamWidthAlongship="7.67" BeamWidthAthwartship="7.96" AngleOffsetAlongship="-0.03" AngleOffsetAthwartship="0.02" />\n <FrequencyPar Frequency="97549" Gain="25.85" Impedance="75" Phase="0" BeamWidthAlongship="8" BeamWidthAthwartship="8.09" AngleOffsetAlongship="0" AngleOffsetAthwartship="0" />\n <FrequencyPar Frequency="98012" Gain="25.85" Impedance="75" Phase="0" BeamWidthAlongship="8.64" BeamWidthAthwartship="8.03" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="98474" Gain="26.07" Impedance="75" Phase="0" BeamWidthAlongship="8.02" BeamWidthAthwartship="8.03" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="98936" Gain="26.16" Impedance="75" Phase="0" BeamWidthAlongship="7.65" BeamWidthAthwartship="7.88" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="99399" Gain="26.01" Impedance="75" Phase="0" BeamWidthAlongship="8" BeamWidthAthwartship="7.8" AngleOffsetAlongship="0.05" AngleOffsetAthwartship="-0.03" />\n <FrequencyPar Frequency="99861" Gain="25.9" Impedance="75" Phase="0" BeamWidthAlongship="8.23" BeamWidthAthwartship="7.81" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.01" />\n <FrequencyPar Frequency="100324" Gain="25.92" Impedance="75" Phase="0" BeamWidthAlongship="7.75" BeamWidthAthwartship="7.77" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="100786" Gain="25.86" Impedance="75" Phase="0" BeamWidthAlongship="7.43" BeamWidthAthwartship="7.6" AngleOffsetAlongship="0.07" AngleOffsetAthwartship="0" />\n <FrequencyPar Frequency="101249" Gain="25.77" Impedance="75" Phase="0" BeamWidthAlongship="7.81" BeamWidthAthwartship="7.63" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="101711" Gain="25.8" Impedance="75" Phase="0" BeamWidthAlongship="7.83" BeamWidthAthwartship="7.69" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="102174" Gain="25.89" Impedance="75" Phase="0" BeamWidthAlongship="7.59" BeamWidthAthwartship="7.66" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="102636" Gain="25.99" Impedance="75" Phase="0" BeamWidthAlongship="7.59" BeamWidthAthwartship="7.59" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.1" />\n <FrequencyPar Frequency="103099" Gain="26.03" Impedance="75" Phase="0" BeamWidthAlongship="7.68" BeamWidthAthwartship="7.6" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="103561" Gain="26.11" Impedance="75" Phase="0" BeamWidthAlongship="7.76" BeamWidthAthwartship="7.62" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="104024" Gain="26.23" Impedance="75" Phase="0" BeamWidthAlongship="7.51" BeamWidthAthwartship="7.46" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.1" />\n <FrequencyPar Frequency="104486" Gain="26.24" Impedance="75" Phase="0" BeamWidthAlongship="7.46" BeamWidthAthwartship="7.43" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="104948" Gain="26.17" Impedance="75" Phase="0" BeamWidthAlongship="7.52" BeamWidthAthwartship="7.4" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="105411" Gain="26.07" Impedance="75" Phase="0" BeamWidthAlongship="7.45" BeamWidthAthwartship="7.36" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="105873" Gain="26.02" Impedance="75" Phase="0" BeamWidthAlongship="7.33" BeamWidthAthwartship="7.32" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="106336" Gain="25.93" Impedance="75" Phase="0" BeamWidthAlongship="7.34" BeamWidthAthwartship="7.28" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="106798" Gain="25.86" Impedance="75" Phase="0" BeamWidthAlongship="7.42" BeamWidthAthwartship="7.3" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="107261" Gain="25.87" Impedance="75" Phase="0" BeamWidthAlongship="7.41" BeamWidthAthwartship="7.31" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="107723" Gain="25.92" Impedance="75" Phase="0" BeamWidthAlongship="7.46" BeamWidthAthwartship="7.3" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="108186" Gain="26.07" Impedance="75" Phase="0" BeamWidthAlongship="7.53" BeamWidthAthwartship="7.27" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="108648" Gain="26.17" Impedance="75" Phase="0" BeamWidthAlongship="7.37" BeamWidthAthwartship="7.29" AngleOffsetAlongship="-0.19" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="109111" Gain="26.29" Impedance="75" Phase="0" BeamWidthAlongship="7.37" BeamWidthAthwartship="7.28" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="109573" Gain="26.35" Impedance="75" Phase="0" BeamWidthAlongship="7.47" BeamWidthAthwartship="7.13" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="110036" Gain="26.38" Impedance="75" Phase="0" BeamWidthAlongship="7.38" BeamWidthAthwartship="7.08" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="110498" Gain="26.38" Impedance="75" Phase="0" BeamWidthAlongship="7.18" BeamWidthAthwartship="7.16" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="110960" Gain="26.26" Impedance="75" Phase="0" BeamWidthAlongship="7.26" BeamWidthAthwartship="7.11" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="111423" Gain="26.21" Impedance="75" Phase="0" BeamWidthAlongship="7.36" BeamWidthAthwartship="7.16" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="111885" Gain="26.16" Impedance="75" Phase="0" BeamWidthAlongship="7.17" BeamWidthAthwartship="7.06" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="112348" Gain="26.23" Impedance="75" Phase="0" BeamWidthAlongship="7.12" BeamWidthAthwartship="7.05" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.1" />\n <FrequencyPar Frequency="112810" Gain="26.25" Impedance="75" Phase="0" BeamWidthAlongship="7.1" BeamWidthAthwartship="7.03" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="113273" Gain="26.45" Impedance="75" Phase="0" BeamWidthAlongship="7.02" BeamWidthAthwartship="7" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="113735" Gain="26.64" Impedance="75" Phase="0" BeamWidthAlongship="6.96" BeamWidthAthwartship="6.9" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="119285" Gain="26.83" Impedance="75" Phase="0" BeamWidthAlongship="6.67" BeamWidthAthwartship="6.63" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="119747" Gain="26.97" Impedance="75" Phase="0" BeamWidthAlongship="6.54" BeamWidthAthwartship="6.5" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="120210" Gain="27.11" Impedance="75" Phase="0" BeamWidthAlongship="6.49" BeamWidthAthwartship="6.49" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="120672" Gain="27.24" Impedance="75" Phase="0" BeamWidthAlongship="6.41" BeamWidthAthwartship="6.42" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="121135" Gain="27.25" Impedance="75" Phase="0" BeamWidthAlongship="6.31" BeamWidthAthwartship="6.34" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="121597" Gain="27.27" Impedance="75" Phase="0" BeamWidthAlongship="6.3" BeamWidthAthwartship="6.34" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="122060" Gain="27.13" Impedance="75" Phase="0" BeamWidthAlongship="6.34" BeamWidthAthwartship="6.27" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="122522" Gain="27.05" Impedance="75" Phase="0" BeamWidthAlongship="6.34" BeamWidthAthwartship="6.27" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="122984" Gain="26.94" Impedance="75" Phase="0" BeamWidthAlongship="6.33" BeamWidthAthwartship="6.25" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="123447" Gain="26.88" Impedance="75" Phase="0" BeamWidthAlongship="6.44" BeamWidthAthwartship="6.23" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="123909" Gain="26.87" Impedance="75" Phase="0" BeamWidthAlongship="6.52" BeamWidthAthwartship="6.23" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="124372" Gain="26.92" Impedance="75" Phase="0" BeamWidthAlongship="6.42" BeamWidthAthwartship="6.27" AngleOffsetAlongship="-0.21" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="124834" Gain="27.13" Impedance="75" Phase="0" BeamWidthAlongship="6.29" BeamWidthAthwartship="6.22" AngleOffsetAlongship="-0.22" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="125297" Gain="27.29" Impedance="75" Phase="0" BeamWidthAlongship="6.31" BeamWidthAthwartship="6.17" AngleOffsetAlongship="-0.21" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="125759" Gain="27.47" Impedance="75" Phase="0" BeamWidthAlongship="6.28" BeamWidthAthwartship="6.14" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="126222" Gain="27.62" Impedance="75" Phase="0" BeamWidthAlongship="6.12" BeamWidthAthwartship="6.08" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.2" />\n <FrequencyPar Frequency="126684" Gain="27.66" Impedance="75" Phase="0" BeamWidthAlongship="6.06" BeamWidthAthwartship="5.99" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="127147" Gain="27.66" Impedance="75" Phase="0" BeamWidthAlongship="6.1" BeamWidthAthwartship="6" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="127609" Gain="27.57" Impedance="75" Phase="0" BeamWidthAlongship="6.14" BeamWidthAthwartship="5.96" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="128072" Gain="27.54" Impedance="75" Phase="0" BeamWidthAlongship="6.04" BeamWidthAthwartship="5.85" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="128534" Gain="27.45" Impedance="75" Phase="0" BeamWidthAlongship="6.09" BeamWidthAthwartship="5.87" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="128996" Gain="27.37" Impedance="75" Phase="0" BeamWidthAlongship="6.21" BeamWidthAthwartship="5.92" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="129459" Gain="27.39" Impedance="75" Phase="0" BeamWidthAlongship="6.17" BeamWidthAthwartship="5.95" AngleOffsetAlongship="-0.19" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="129921" Gain="27.52" Impedance="75" Phase="0" BeamWidthAlongship="6.01" BeamWidthAthwartship="6" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.21" />\n <FrequencyPar Frequency="130384" Gain="27.67" Impedance="75" Phase="0" BeamWidthAlongship="6.03" BeamWidthAthwartship="5.94" AngleOffsetAlongship="-0.21" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="130846" Gain="27.87" Impedance="75" Phase="0" BeamWidthAlongship="6.04" BeamWidthAthwartship="5.87" AngleOffsetAlongship="-0.22" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="131309" Gain="28.01" Impedance="75" Phase="0" BeamWidthAlongship="5.86" BeamWidthAthwartship="5.79" AngleOffsetAlongship="-0.21" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="131771" Gain="28.12" Impedance="75" Phase="0" BeamWidthAlongship="5.8" BeamWidthAthwartship="5.83" AngleOffsetAlongship="-0.21" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="132234" Gain="28.08" Impedance="75" Phase="0" BeamWidthAlongship="5.84" BeamWidthAthwartship="5.78" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="132696" Gain="28" Impedance="75" Phase="0" BeamWidthAlongship="5.83" BeamWidthAthwartship="5.68" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="133159" Gain="27.86" Impedance="75" Phase="0" BeamWidthAlongship="5.88" BeamWidthAthwartship="5.66" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="133621" Gain="27.52" Impedance="75" Phase="0" BeamWidthAlongship="5.83" BeamWidthAthwartship="5.72" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="140096" Gain="26.98" Impedance="75" Phase="0" BeamWidthAlongship="5.5" BeamWidthAthwartship="5.39" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="140558" Gain="26.93" Impedance="75" Phase="0" BeamWidthAlongship="5.45" BeamWidthAthwartship="5.42" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="141021" Gain="26.93" Impedance="75" Phase="0" BeamWidthAlongship="5.52" BeamWidthAthwartship="5.36" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="141483" Gain="27.01" Impedance="75" Phase="0" BeamWidthAlongship="5.45" BeamWidthAthwartship="5.32" AngleOffsetAlongship="-0.19" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="141945" Gain="27.16" Impedance="75" Phase="0" BeamWidthAlongship="5.36" BeamWidthAthwartship="5.33" AngleOffsetAlongship="-0.21" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="142408" Gain="27.39" Impedance="75" Phase="0" BeamWidthAlongship="5.3" BeamWidthAthwartship="5.28" AngleOffsetAlongship="-0.21" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="142870" Gain="27.67" Impedance="75" Phase="0" BeamWidthAlongship="5.29" BeamWidthAthwartship="5.26" AngleOffsetAlongship="-0.19" AngleOffsetAthwartship="0.2" />\n <FrequencyPar Frequency="143333" Gain="27.97" Impedance="75" Phase="0" BeamWidthAlongship="5.31" BeamWidthAthwartship="5.18" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="143795" Gain="28.13" Impedance="75" Phase="0" BeamWidthAlongship="5.24" BeamWidthAthwartship="5.19" AngleOffsetAlongship="-0.19" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="144258" Gain="28.09" Impedance="75" Phase="0" BeamWidthAlongship="5.25" BeamWidthAthwartship="5.19" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="144720" Gain="27.98" Impedance="75" Phase="0" BeamWidthAlongship="5.25" BeamWidthAthwartship="5.17" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="145183" Gain="27.86" Impedance="75" Phase="0" BeamWidthAlongship="5.19" BeamWidthAthwartship="5.15" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="145645" Gain="27.77" Impedance="75" Phase="0" BeamWidthAlongship="5.2" BeamWidthAthwartship="5.07" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="146108" Gain="27.75" Impedance="75" Phase="0" BeamWidthAlongship="5.16" BeamWidthAthwartship="5" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="146570" Gain="27.71" Impedance="75" Phase="0" BeamWidthAlongship="5.13" BeamWidthAthwartship="5.07" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="147033" Gain="27.74" Impedance="75" Phase="0" BeamWidthAlongship="5.07" BeamWidthAthwartship="5.03" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="147495" Gain="27.8" Impedance="75" Phase="0" BeamWidthAlongship="4.95" BeamWidthAthwartship="5.01" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="147957" Gain="27.86" Impedance="75" Phase="0" BeamWidthAlongship="4.95" BeamWidthAthwartship="4.96" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="148420" Gain="27.96" Impedance="75" Phase="0" BeamWidthAlongship="4.97" BeamWidthAthwartship="4.92" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="148882" Gain="28" Impedance="75" Phase="0" BeamWidthAlongship="4.95" BeamWidthAthwartship="4.92" AngleOffsetAlongship="-0.19" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="149345" Gain="28.04" Impedance="75" Phase="0" BeamWidthAlongship="4.9" BeamWidthAthwartship="4.91" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="149807" Gain="28" Impedance="75" Phase="0" BeamWidthAlongship="4.92" BeamWidthAthwartship="4.88" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="150270" Gain="27.89" Impedance="75" Phase="0" BeamWidthAlongship="4.89" BeamWidthAthwartship="4.88" AngleOffsetAlongship="-0.14" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="150732" Gain="27.78" Impedance="75" Phase="0" BeamWidthAlongship="4.84" BeamWidthAthwartship="4.89" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="151195" Gain="27.74" Impedance="75" Phase="0" BeamWidthAlongship="4.81" BeamWidthAthwartship="4.78" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="151657" Gain="27.67" Impedance="75" Phase="0" BeamWidthAlongship="4.84" BeamWidthAthwartship="4.79" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="152120" Gain="27.68" Impedance="75" Phase="0" BeamWidthAlongship="4.84" BeamWidthAthwartship="4.79" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="152582" Gain="27.75" Impedance="75" Phase="0" BeamWidthAlongship="4.79" BeamWidthAthwartship="4.85" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.2" />\n <FrequencyPar Frequency="153045" Gain="27.88" Impedance="75" Phase="0" BeamWidthAlongship="4.8" BeamWidthAthwartship="4.82" AngleOffsetAlongship="-0.19" AngleOffsetAthwartship="0.23" />\n <FrequencyPar Frequency="153507" Gain="28.02" Impedance="75" Phase="0" BeamWidthAlongship="4.79" BeamWidthAthwartship="4.8" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.2" />\n <FrequencyPar Frequency="153969" Gain="28.17" Impedance="75" Phase="0" BeamWidthAlongship="4.74" BeamWidthAthwartship="4.78" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.21" />\n <FrequencyPar Frequency="154432" Gain="28.32" Impedance="75" Phase="0" BeamWidthAlongship="4.71" BeamWidthAthwartship="4.74" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="154894" Gain="28.47" Impedance="75" Phase="0" BeamWidthAlongship="4.73" BeamWidthAthwartship="4.72" AngleOffsetAlongship="-0.18" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="155357" Gain="28.55" Impedance="75" Phase="0" BeamWidthAlongship="4.7" BeamWidthAthwartship="4.78" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="155819" Gain="28.66" Impedance="75" Phase="0" BeamWidthAlongship="4.71" BeamWidthAthwartship="4.7" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="156282" Gain="28.71" Impedance="75" Phase="0" BeamWidthAlongship="4.67" BeamWidthAthwartship="4.75" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="156744" Gain="28.66" Impedance="75" Phase="0" BeamWidthAlongship="4.67" BeamWidthAthwartship="4.71" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="157207" Gain="28.54" Impedance="75" Phase="0" BeamWidthAlongship="4.69" BeamWidthAthwartship="4.67" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="157669" Gain="28.36" Impedance="75" Phase="0" BeamWidthAlongship="4.54" BeamWidthAthwartship="4.75" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="158000" Gain="28.29" Impedance="75" Phase="0" BeamWidthAlongship="4.54" BeamWidthAthwartship="4.66" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.15" />\n </Transducer>\n </Channel>\n </Channels>\n </Transceiver>\n <Transceiver TransceiverName="WBT 400145" IPAddress="157.237.15.105" MarketSegment="Scientific" SerialNumber="400145" Impedance="5400" Multiplexing="0" RxSampleFrequency="1500000" TransceiverType="WBT" EthernetAddress="009072061b11" Version="[0] Ethernet: 00:90:72:06:1B:11
[1] Parts-list: WBT 371790/F
[2] Product: WBT
IP Address: 157.237.15.105
Subnet mask: 255.255.0.0
Default gateway: 157.237.15.1
Serial number: 400145
Embedded software: Rev. 2.54
FPGA TX firmware: Rev. 5
FPGA RX firmware: Rev. 7
CH1: 518W CH2: 515W CH3: 510W CH4: 509W
TRD1: Unable to detect transducer
TRD2: Unable to detect transducer
TRD3: Unable to detect transducer
TRD4: Unable to detect transducer
" TransceiverSoftwareVersion="2.54" TransceiverNumber="5">\n <Channels>\n <Channel ChannelID="WBT 400145-15 ES200-7C_ES" LogicalChannelID="WBT 400145-15 ES200-7C" ChannelIdShort="ES200 Shimada" MaxTxPowerTransceiver="2000" HWChannelConfiguration="15" PulseDuration="6.4E-05;0.000128;0.000256;0.000512;0.001024" PulseDurationFM="0.000512;0.001024;0.002048;0.004096;0.008192">\n <Transducer TransducerName="ES200-7C" ArticleNumber="KSV-203003" SerialNumber="0" Frequency="200000" FrequencyMinimum="160000" FrequencyMaximum="260000" MaxTxPowerTransducer="150" Gain="26;26;26;26;27.55" SaCorrection="0;0;0;0;-0.09" EquivalentBeamAngle="-20.7" DirectivityDropAt2XBeamWidth="0" AngleSensitivityAlongship="23" AngleSensitivityAthwartship="23" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.25" BeamWidthAlongship="6.79" BeamWidthAthwartship="6.07" BeamType="1">\n <FrequencyPar Frequency="162000" Gain="24.16" Impedance="75" Phase="0" BeamWidthAlongship="11.32" BeamWidthAthwartship="7.77" AngleOffsetAlongship="0.9" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="162981" Gain="24.84" Impedance="75" Phase="0" BeamWidthAlongship="9.04" BeamWidthAthwartship="7.06" AngleOffsetAlongship="0.17" AngleOffsetAthwartship="0.1" />\n <FrequencyPar Frequency="163962" Gain="25.38" Impedance="75" Phase="0" BeamWidthAlongship="10.95" BeamWidthAthwartship="7.62" AngleOffsetAlongship="0.22" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="164943" Gain="28.08" Impedance="75" Phase="0" BeamWidthAlongship="8.84" BeamWidthAthwartship="7.8" AngleOffsetAlongship="-0.04" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="165924" Gain="24.69" Impedance="75" Phase="0" BeamWidthAlongship="8.95" BeamWidthAthwartship="7.79" AngleOffsetAlongship="-0.11" AngleOffsetAthwartship="0.2" />\n <FrequencyPar Frequency="166905" Gain="25.39" Impedance="75" Phase="0" BeamWidthAlongship="8.15" BeamWidthAthwartship="7.63" AngleOffsetAlongship="-0.22" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="167886" Gain="25.48" Impedance="75" Phase="0" BeamWidthAlongship="8.08" BeamWidthAthwartship="7.1" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="168867" Gain="25.8" Impedance="75" Phase="0" BeamWidthAlongship="8.66" BeamWidthAthwartship="7.52" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="169848" Gain="26.26" Impedance="75" Phase="0" BeamWidthAlongship="8.2" BeamWidthAthwartship="7.14" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="179658" Gain="26.42" Impedance="75" Phase="0" BeamWidthAlongship="8.82" BeamWidthAthwartship="7.74" AngleOffsetAlongship="-0.03" AngleOffsetAthwartship="0.1" />\n <FrequencyPar Frequency="180639" Gain="26.71" Impedance="75" Phase="0" BeamWidthAlongship="8.73" BeamWidthAthwartship="8.09" AngleOffsetAlongship="0.05" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="181620" Gain="27.17" Impedance="75" Phase="0" BeamWidthAlongship="8.15" BeamWidthAthwartship="7.69" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="182601" Gain="27.17" Impedance="75" Phase="0" BeamWidthAlongship="8.46" BeamWidthAthwartship="7.88" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="183582" Gain="26.95" Impedance="75" Phase="0" BeamWidthAlongship="8.12" BeamWidthAthwartship="7.45" AngleOffsetAlongship="-0.06" AngleOffsetAthwartship="0.07" />\n <FrequencyPar Frequency="184563" Gain="26.61" Impedance="75" Phase="0" BeamWidthAlongship="8.79" BeamWidthAthwartship="7.82" AngleOffsetAlongship="-0.03" AngleOffsetAthwartship="0.05" />\n <FrequencyPar Frequency="185544" Gain="26.83" Impedance="75" Phase="0" BeamWidthAlongship="8.15" BeamWidthAthwartship="7.15" AngleOffsetAlongship="0.03" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="186525" Gain="27.31" Impedance="75" Phase="0" BeamWidthAlongship="8.15" BeamWidthAthwartship="7.16" AngleOffsetAlongship="0.03" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="187506" Gain="27.62" Impedance="75" Phase="0" BeamWidthAlongship="7.74" BeamWidthAthwartship="7.2" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="188486" Gain="27.57" Impedance="75" Phase="0" BeamWidthAlongship="7.91" BeamWidthAthwartship="7.33" AngleOffsetAlongship="-0.08" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="189467" Gain="27.29" Impedance="75" Phase="0" BeamWidthAlongship="7.59" BeamWidthAthwartship="7.21" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="190448" Gain="27.07" Impedance="75" Phase="0" BeamWidthAlongship="8.13" BeamWidthAthwartship="7.25" AngleOffsetAlongship="0.01" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="191429" Gain="27.44" Impedance="75" Phase="0" BeamWidthAlongship="7.38" BeamWidthAthwartship="6.6" AngleOffsetAlongship="0.01" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="192410" Gain="27.7" Impedance="75" Phase="0" BeamWidthAlongship="7.62" BeamWidthAthwartship="7.05" AngleOffsetAlongship="-0.01" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="193391" Gain="27.83" Impedance="75" Phase="0" BeamWidthAlongship="7.38" BeamWidthAthwartship="6.88" AngleOffsetAlongship="-0.15" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="194372" Gain="27.66" Impedance="75" Phase="0" BeamWidthAlongship="7.35" BeamWidthAthwartship="6.75" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="195353" Gain="27.31" Impedance="75" Phase="0" BeamWidthAlongship="7.03" BeamWidthAthwartship="6.89" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="196334" Gain="27.31" Impedance="75" Phase="0" BeamWidthAlongship="7.15" BeamWidthAthwartship="6.5" AngleOffsetAlongship="-0.05" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="197315" Gain="27.66" Impedance="75" Phase="0" BeamWidthAlongship="7.02" BeamWidthAthwartship="6.58" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="198296" Gain="27.98" Impedance="75" Phase="0" BeamWidthAlongship="6.87" BeamWidthAthwartship="6.45" AngleOffsetAlongship="-0.1" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="199277" Gain="27.99" Impedance="75" Phase="0" BeamWidthAlongship="7.23" BeamWidthAthwartship="6.43" AngleOffsetAlongship="-0.16" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="200258" Gain="27.73" Impedance="75" Phase="0" BeamWidthAlongship="7.28" BeamWidthAthwartship="6.57" AngleOffsetAlongship="-0.22" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="201239" Gain="27.53" Impedance="75" Phase="0" BeamWidthAlongship="7.05" BeamWidthAthwartship="6.32" AngleOffsetAlongship="-0.2" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="202220" Gain="27.72" Impedance="75" Phase="0" BeamWidthAlongship="7.03" BeamWidthAthwartship="6.31" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="203201" Gain="28.02" Impedance="75" Phase="0" BeamWidthAlongship="6.77" BeamWidthAthwartship="6.44" AngleOffsetAlongship="0.07" AngleOffsetAthwartship="0.15" />\n <FrequencyPar Frequency="204182" Gain="28.29" Impedance="75" Phase="0" BeamWidthAlongship="7.39" BeamWidthAthwartship="6.28" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.1" />\n <FrequencyPar Frequency="205163" Gain="28.13" Impedance="75" Phase="0" BeamWidthAlongship="7.32" BeamWidthAthwartship="6.73" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="206144" Gain="27.84" Impedance="75" Phase="0" BeamWidthAlongship="7.03" BeamWidthAthwartship="6.35" AngleOffsetAlongship="-0.17" AngleOffsetAthwartship="0.04" />\n <FrequencyPar Frequency="215954" Gain="28.94" Impedance="75" Phase="0" BeamWidthAlongship="6.61" BeamWidthAthwartship="6.01" AngleOffsetAlongship="0.05" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="216935" Gain="28.81" Impedance="75" Phase="0" BeamWidthAlongship="6.67" BeamWidthAthwartship="6.17" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="217916" Gain="28.46" Impedance="75" Phase="0" BeamWidthAlongship="6.51" BeamWidthAthwartship="6.11" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="218897" Gain="28.19" Impedance="75" Phase="0" BeamWidthAlongship="6.81" BeamWidthAthwartship="5.8" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.17" />\n <FrequencyPar Frequency="219878" Gain="28.44" Impedance="75" Phase="0" BeamWidthAlongship="6.46" BeamWidthAthwartship="5.9" AngleOffsetAlongship="0.08" AngleOffsetAthwartship="0.2" />\n <FrequencyPar Frequency="220859" Gain="28.89" Impedance="75" Phase="0" BeamWidthAlongship="6.3" BeamWidthAthwartship="5.74" AngleOffsetAlongship="0.07" AngleOffsetAthwartship="0.19" />\n <FrequencyPar Frequency="221840" Gain="29.07" Impedance="75" Phase="0" BeamWidthAlongship="6.24" BeamWidthAthwartship="5.88" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="222821" Gain="28.78" Impedance="75" Phase="0" BeamWidthAlongship="6.7" BeamWidthAthwartship="6.13" AngleOffsetAlongship="0.03" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="223802" Gain="27.5" Impedance="75" Phase="0" BeamWidthAlongship="6.33" BeamWidthAthwartship="6.05" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.16" />\n <FrequencyPar Frequency="224783" Gain="26.81" Impedance="75" Phase="0" BeamWidthAlongship="6.37" BeamWidthAthwartship="6.02" AngleOffsetAlongship="0.04" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="225764" Gain="27.19" Impedance="75" Phase="0" BeamWidthAlongship="6.02" BeamWidthAthwartship="5.78" AngleOffsetAlongship="0.02" AngleOffsetAthwartship="0.12" />\n <FrequencyPar Frequency="226745" Gain="27.7" Impedance="75" Phase="0" BeamWidthAlongship="7.76" BeamWidthAthwartship="5.68" AngleOffsetAlongship="0.17" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="227726" Gain="28.74" Impedance="75" Phase="0" BeamWidthAlongship="6.17" BeamWidthAthwartship="5.83" AngleOffsetAlongship="-0.04" AngleOffsetAthwartship="0.18" />\n <FrequencyPar Frequency="228707" Gain="28.36" Impedance="75" Phase="0" BeamWidthAlongship="6.35" BeamWidthAthwartship="5.95" AngleOffsetAlongship="-0.13" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="229688" Gain="27.99" Impedance="75" Phase="0" BeamWidthAlongship="6.1" BeamWidthAthwartship="5.69" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="230669" Gain="28.06" Impedance="75" Phase="0" BeamWidthAlongship="6.22" BeamWidthAthwartship="5.86" AngleOffsetAlongship="0.12" AngleOffsetAthwartship="0.06" />\n <FrequencyPar Frequency="231650" Gain="28.44" Impedance="75" Phase="0" BeamWidthAlongship="6.06" BeamWidthAthwartship="5.8" AngleOffsetAlongship="0.04" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="232631" Gain="28.68" Impedance="75" Phase="0" BeamWidthAlongship="6.15" BeamWidthAthwartship="5.67" AngleOffsetAlongship="-0.01" AngleOffsetAthwartship="0.14" />\n <FrequencyPar Frequency="233612" Gain="28.53" Impedance="75" Phase="0" BeamWidthAlongship="6.24" BeamWidthAthwartship="5.71" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="234593" Gain="27.93" Impedance="75" Phase="0" BeamWidthAlongship="5.78" BeamWidthAthwartship="6.11" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="235574" Gain="27.9" Impedance="75" Phase="0" BeamWidthAlongship="6.6" BeamWidthAthwartship="5.77" AngleOffsetAlongship="-0.09" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="236555" Gain="28.31" Impedance="75" Phase="0" BeamWidthAlongship="6.15" BeamWidthAthwartship="5.55" AngleOffsetAlongship="0" AngleOffsetAthwartship="0.13" />\n <FrequencyPar Frequency="237536" Gain="28.77" Impedance="75" Phase="0" BeamWidthAlongship="5.98" BeamWidthAthwartship="5.49" AngleOffsetAlongship="-0.02" AngleOffsetAthwartship="0.09" />\n <FrequencyPar Frequency="238517" Gain="28.81" Impedance="75" Phase="0" BeamWidthAlongship="6.6" BeamWidthAthwartship="5.29" AngleOffsetAlongship="-0.25" AngleOffsetAthwartship="0.03" />\n <FrequencyPar Frequency="239497" Gain="28.41" Impedance="75" Phase="0" BeamWidthAlongship="6.25" BeamWidthAthwartship="5.91" AngleOffsetAlongship="-0.12" AngleOffsetAthwartship="0.11" />\n <FrequencyPar Frequency="240478" Gain="28.33" Impedance="75" Phase="0" BeamWidthAlongship="5.95" BeamWidthAthwartship="5.55" AngleOffsetAlongship="-0.07" AngleOffsetAthwartship="0.08" />\n <FrequencyPar Frequency="241459" Gain="28.35" Impedance="75" Phase="0" BeamWidthAlongship="5.85" BeamWidthAthwartship="5.38" AngleOffsetAlongship="0.02" AngleOffsetAthwartship="0.09" />\n </Transducer>\n </Channel>\n </Channels>\n </Transceiver>\n </Transceivers>\n <Transducers MergeOperation="AddNodeTree">\n <Transducer TransducerName="ES18" TransducerSerialNumber="0" TransducerCustomName="ES18 Shimada" TransducerMounting="DropKeel" TransducerOrientation="Vertical" TransducerOffsetX="0" TransducerOffsetY="0" TransducerOffsetZ="0" TransducerAlphaX="0" TransducerAlphaY="0" TransducerAlphaZ="0" />\n <Transducer TransducerName="ES38B" TransducerSerialNumber="0" TransducerCustomName="ES 38 Shimada" TransducerMounting="DropKeel" TransducerOrientation="Vertical" TransducerOffsetX="0" TransducerOffsetY="0" TransducerOffsetZ="0" TransducerAlphaX="0" TransducerAlphaY="0" TransducerAlphaZ="0" />\n <Transducer TransducerName="ES70-7C" TransducerSerialNumber="0" TransducerCustomName="ES70 Shimada" TransducerMounting="DropKeel" TransducerOrientation="Vertical" TransducerOffsetX="0" TransducerOffsetY="0" TransducerOffsetZ="0" TransducerAlphaX="0" TransducerAlphaY="0" TransducerAlphaZ="0" />\n <Transducer TransducerName="ES120-7C" TransducerSerialNumber="0" TransducerCustomName="ES120 Shimada" TransducerMounting="DropKeel" TransducerOrientation="Vertical" TransducerOffsetX="0" TransducerOffsetY="0" TransducerOffsetZ="0" TransducerAlphaX="0" TransducerAlphaY="0" TransducerAlphaZ="0" />\n <Transducer TransducerName="ES200-7C" TransducerSerialNumber="0" TransducerCustomName="ES200 Shimada" TransducerMounting="DropKeel" TransducerOrientation="Vertical" TransducerOffsetX="0" TransducerOffsetY="0" TransducerOffsetZ="0" TransducerAlphaX="0" TransducerAlphaY="0" TransducerAlphaZ="0" />\n </Transducers>\n <ConfiguredSensors MergeOperation="AddNodeTree">\n <Sensor Name=" Furuno GPS From Serial Port 8" Type="GPS" Port="Serial Port 8" TalkerID="" X="0" Y="0" Z="0" AngleX="0" AngleY="0" AngleZ="0" Unique="0" Timeout="20">\n <Telegram Name="GLL from Furuno GPS From Serial Port 8" SensorType="GPS" Type="GLL" SubscriptionPath=" Furuno GPS From Serial Port 8@GPS.Geographical.Position" Enabled="1">\n <Value Name="Latitude" Priority="3" />\n <Value Name="Longitude" Priority="3" />\n </Telegram>\n <Telegram Name="GGA from Furuno GPS From Serial Port 8" SensorType="GPS" Type="GGA" SubscriptionPath=" Furuno GPS From Serial Port 8@GPS.Global.Position" Enabled="1">\n <Value Name="Latitude" Priority="4" />\n <Value Name="Longitude" Priority="4" />\n </Telegram>\n <Telegram Name="RMC from Furuno GPS From Serial Port 8" SensorType="GPS" Type="RMC" SubscriptionPath=" Furuno GPS From Serial Port 8@GPS.Specific.PositionSpeedCourse" Enabled="1">\n <Value Name="Latitude" Priority="5" />\n <Value Name="Longitude" Priority="5" />\n <Value Name="Course" Priority="7" />\n <Value Name="Speed" Priority="3" />\n </Telegram>\n <Telegram Name="VTG from Furuno GPS From Serial Port 8" SensorType="GPS" Type="VTG" SubscriptionPath=" Furuno GPS From Serial Port 8@GPS.Ground" Enabled="1">\n <Value Name="Course" Priority="8" />\n <Value Name="CourseNotUsedMagnetic" Priority="1" />\n <Value Name="Speed" Priority="4" />\n </Telegram>\n <Telegram Name="ZDA from Furuno GPS From Serial Port 8" SensorType="GPS" Type="ZDA" SubscriptionPath=" Furuno GPS From Serial Port 8@GPS.TimeInfo" Enabled="1">\n <Value Name="TimeInfo" Priority="1" />\n </Telegram>\n </Sensor>\n <Sensor Name="Gyro From POS/MV Serial Port 13" Type="Gyro" Port="Serial Port 13" TalkerID="" X="0" Y="0" Z="0" AngleX="0" AngleY="0" AngleZ="0" Unique="0" Timeout="20">\n <Telegram Name="HDT from Gyro From POS/MV Serial Port 13" SensorType="Gyro" Type="HDT" SubscriptionPath="Gyro From POS/MV Serial Port 13@Gyro.HeadingTrue" Enabled="1">\n <Value Name="Heading" Priority="1" />\n </Telegram>\n <Telegram Name="HDM from Gyro From POS/MV Serial Port 13" SensorType="Gyro" Type="HDM" SubscriptionPath="Gyro From POS/MV Serial Port 13@Gyro.HeadingMagnetic" Enabled="1">\n <Value Name="Heading" Priority="2" />\n </Telegram>\n <Telegram Name="HDG from Gyro From POS/MV Serial Port 13" SensorType="Gyro" Type="HDG" SubscriptionPath="Gyro From POS/MV Serial Port 13@Gyro.HeadingMagneticDeviation" Enabled="1">\n <Value Name="Heading" Priority="4" />\n </Telegram>\n <Telegram Name="THS from Gyro From POS/MV Serial Port 13" SensorType="Gyro" Type="THS" SubscriptionPath="Gyro From POS/MV Serial Port 13@Gyro.HeadingTrueStatus" Enabled="1">\n <Value Name="Heading" Priority="6" />\n </Telegram>\n </Sensor>\n <Sensor Name="ITI-FS From FS70" Type="ITI-FS" Port="Serial Port 11" TalkerID="II" X="0" Y="0" Z="0" AngleX="0" AngleY="0" AngleZ="0" Unique="0" Timeout="20">\n <Telegram Name="ITI-FS Datagrams from ITI-FS From FS70" SensorType="ITI-FS" Type="ITI-FS Datagrams" SubscriptionPath="ITI-FS From FS70@ITI-FS.ITI-FS" Enabled="1">\n <Value Name="ITI-FS" Priority="2" />\n </Telegram>\n </Sensor>\n <Sensor Name="ITI-FS From Serial Port 9" Type="ITI-FS" Port="Serial Port 9" TalkerID="II" X="0" Y="0" Z="0" AngleX="0" AngleY="0" AngleZ="0" Unique="0" Timeout="20">\n <Telegram Name="ITI-FS Datagrams from ITI-FS From Serial Port 9" SensorType="ITI-FS" Type="ITI-FS Datagrams" SubscriptionPath="ITI-FS From Serial Port 9@ITI-FS.ITI-FS" Enabled="1">\n <Value Name="ITI-FS" Priority="2" />\n </Telegram>\n </Sensor>\n <Sensor Name="GPS From POS/MV Port 13" Type="GPS" Port="Serial Port 13" TalkerID="" X="0" Y="0" Z="0" AngleX="0" AngleY="0" AngleZ="0" Unique="0" Timeout="20">\n <Telegram Name="GGA from GPS From POS/MV Port 13" SensorType="GPS" Type="GGA" SubscriptionPath="GPS From POS/MV Port 13@GPS.Global.Position" Enabled="1">\n <Value Name="Latitude" Priority="1" />\n <Value Name="Longitude" Priority="1" />\n </Telegram>\n <Telegram Name="RMC from GPS From POS/MV Port 13" SensorType="GPS" Type="RMC" SubscriptionPath="GPS From POS/MV Port 13@GPS.Specific.PositionSpeedCourse" Enabled="1">\n <Value Name="Latitude" Priority="2" />\n <Value Name="Longitude" Priority="2" />\n <Value Name="Course" Priority="3" />\n <Value Name="Speed" Priority="1" />\n </Telegram>\n <Telegram Name="VTG from GPS From POS/MV Port 13" SensorType="GPS" Type="VTG" SubscriptionPath="GPS From POS/MV Port 13@GPS.Ground" Enabled="1">\n <Value Name="Course" Priority="5" />\n <Value Name="CourseNotUsedMagnetic" Priority="2" />\n <Value Name="Speed" Priority="2" />\n </Telegram>\n <Telegram Name="ZDA from GPS From POS/MV Port 13" SensorType="GPS" Type="ZDA" SubscriptionPath="GPS From POS/MV Port 13@GPS.TimeInfo" Enabled="1">\n <Value Name="TimeInfo" Priority="2" />\n </Telegram>\n </Sensor>\n <Sensor Name="Motion EM3000 From Serial Port 12" Type="MotionBinary" Port="Serial Port 12" TalkerID="" X="0" Y="0" Z="0" AngleX="0" AngleY="0" AngleZ="0" Unique="0" Timeout="20">\n <Telegram Name="MRU from Motion EM3000 From Serial Port 12" SensorType="MotionBinary" Type="MRU" SubscriptionPath="Motion EM3000 From Serial Port 12@MotionBinary.MotionData" Enabled="1">\n <Value Name="Roll" Priority="2" />\n <Value Name="Pitch" Priority="2" />\n <Value Name="Heading" Priority="9" />\n <Value Name="Heave" Priority="2" />\n </Telegram>\n </Sensor>\n <Sensor Name="Motion TSS1 From Serial Port 10" Type="MotionAscii" Port="Serial Port 10" TalkerID="" X="0" Y="0" Z="0" AngleX="0" AngleY="0" AngleZ="0" Unique="0" Timeout="20">\n <Telegram Name="MRU from Motion TSS1 From Serial Port 10" SensorType="MotionAscii" Type="MRU" SubscriptionPath="Motion TSS1 From Serial Port 10@MotionAscii.MotionData" Enabled="1">\n <Value Name="Roll" Priority="1" />\n <Value Name="Pitch" Priority="1" />\n <Value Name="Heading_Dummy" Priority="1" />\n <Value Name="Heave" Priority="1" />\n </Telegram>\n </Sensor>\n </ConfiguredSensors>\n</Configuration>', dtype='<U74340')
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - pulse_length_binPandasIndex
PandasIndex(Index([0, 1, 2, 3, 4], dtype='int64', name='pulse_length_bin'))
- cal_frequencyPandasIndex
PandasIndex(Index([ 45000, 45473, 45947, 46420, 46894, 47367, 47841, 48314, 48788, 49261, ... 232631, 233612, 234593, 235574, 236555, 237536, 238517, 239497, 240478, 241459], dtype='int64', name='cal_frequency', length=269)) - cal_channel_idPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='cal_channel_id'))
<xarray.DatasetView> Size: 353kB Dimensions: (channel: 5, pulse_length_bin: 5, cal_channel_id: 3, cal_frequency: 269, WBT_filter_n: 47, PC_filter_n: 123) Coordinates: * channel (channel) <U25 500B 'WBT 400140-15 ES120-7C_... * pulse_length_bin (pulse_length_bin) int64 40B 0 1 2 3 4 * cal_frequency (cal_frequency) int64 2kB 45000 ... 241459 * cal_channel_id (cal_channel_id) object 24B 'WBT 400140-15 E... Dimensions without coordinates: WBT_filter_n, PC_filter_n Data variables: (12/21) frequency_nominal (channel) float64 40B 1.2e+05 1.8e+04 ... 2e+05 sa_correction (channel, pulse_length_bin) float64 200B 0.0... gain_correction (channel, pulse_length_bin) float64 200B 27.... pulse_length (channel, pulse_length_bin) float64 200B 6.4... impedance_transceiver (channel) int64 40B 5400 5400 5400 5400 5400 receiver_sampling_frequency (channel) float64 40B 1.5e+06 ... 1.5e+06 ... ... WBT_filter_r (channel, WBT_filter_n) float32 940B 0.00045... WBT_decimation (channel) int64 40B 6 6 6 6 6 PC_filter_i (channel, PC_filter_n) float32 2kB -2.988e-0... PC_filter_r (channel, PC_filter_n) float32 2kB -5.649e-0... PC_decimation (channel) int64 40B 10 7 12 10 8 config_xml <U74340 297kB '<?xml version="1.0" encoding=...xarray.DatasetView
There are a few things to note in this example: - All backscatter data are in the Sonar/Beam_group1 group - The backscatter data and many other variables are aligned in 3 major dimensions: channel, ping_time, range_sample - channel includes all the transducer channel, and for EK80 files these are the channel IDs configured in the EK80 system during data collection. - We chose to use channel instead of frequency to align the data, because some setups use multiple transducers transmitting at the same frequency but pointing in different directions. Additionally, “frequency” can be a misnomer broadband transmissions - We store the “frequency” of the channels in the variable frequency_nominal for easy access - range_sample are 0-based sample count of the received echoes - Useful overview type of information are stored in the Top-level group, which is the “root” of the netCDF file - Raw NMEA datagrams are found in the Platform/NMEA group - Raw CONFIG datagram, which is an XML string, is save in the variable content in the Vendor_specific group
In addition, since ed is just converted and has not been saved, everything is stored in-memory, as indicated in the first line printed out below.
Echopype does not yet support ping-sequence files! We expect that to be part of an upcoming releases.
Saving the EchoData object
Echopype supports saving the EchoData object into a netCDF file (.nc) or a zarr store (.zarr). The groups and the representation stay the same, just the format specification is different. NetCDF files are single files that are easy to handle on a conventional filesystem. Zarr stores are optimized for cloud storage, but can be messy to access on a local filesystem as it would show up as many many folder. Both formats and the tools to access them continue to develop, and in particular there are recent new packages that allow representing multiple netCDF files as a virtual zarr store, so things are becoming more convenient and powerful on the user side!
Lazy-load an EchoData object
We can load the saved zarr back using the open_converted function.
By using chunks={}, we “lazy-load” all the data in this zarr file into a new EchoData object, ed_new, which can be seen as a representation or a handle of the data stored on disk.
This capability is very important for distributed computing and is key to out-of-core computation, because different “workers” can directly access different parts of the same file on disk, and don’t have load the entire dataset into memory. This file is pretty small, but in the OOI_eclipse.ipynb notebook, you can see an example in which the zarr file is approximately 8 GB and its computation would generally overwhelm a regular laptop without distributed computing capability.
We can find out if a variable is lazy-loaded or completely loaded into memory by looking at the representation:
<xarray.DataArray 'backscatter_r' (channel: 5, ping_time: 213,
range_sample: 36198)> Size: 154MB
array([[[ 10.383183 , 13.875601 , 16.015736 , ...,
nan, nan, nan],
[ 10.430219 , 13.910878 , 16.051014 , ...,
nan, nan, nan],
[ 10.383183 , 13.875601 , 16.015736 , ...,
nan, nan, nan],
...,
[ 10.465496 , 13.969673 , 16.121567 , ...,
nan, nan, nan],
[ 10.465496 , 13.969673 , 16.121567 , ...,
nan, nan, nan],
[ 10.430219 , 13.899119 , 16.051014 , ...,
nan, nan, nan]],
[[ -11.782502 , -5.7031074 , -0.50563633, ...,
-125.868164 , -126.09158 , -126.29149 ],
[ -11.711948 , -5.6443124 , -0.47035936, ...,
-133.06467 , -130.23074 , -127.82016 ],
[ -11.770743 , -5.6795893 , -0.49387732, ...,
-133.14697 , -132.48848 , -131.8182 ],
...
[ 3.5394542 , 10.41846 , 15.780557 , ...,
nan, nan, nan],
[ 3.4689002 , 10.383183 , 15.757039 , ...,
nan, nan, nan],
[ 3.4924183 , 10.383183 , 15.757039 , ...,
nan, nan, nan]],
[[ 6.067636 , 8.866274 , 10.747711 , ...,
nan, nan, nan],
[ 6.079395 , 8.842756 , 10.747711 , ...,
nan, nan, nan],
[ 6.2087436 , 9.019141 , 10.912337 , ...,
nan, nan, nan],
...,
[ 6.3028154 , 9.077936 , 10.959373 , ...,
nan, nan, nan],
[ 6.2087436 , 9.0309 , 10.935855 , ...,
nan, nan, nan],
[ 6.3028154 , 9.077936 , 10.971132 , ...,
nan, nan, nan]]], dtype=float32)
Coordinates:
* channel (channel) <U25 500B 'WBT 400140-15 ES120-7C_ES' ... 'WBT 40...
* ping_time (ping_time) datetime64[ns] 2kB 2023-08-11T16:57:27.277163 ....
* range_sample (range_sample) int64 290kB 0 1 2 3 ... 36194 36195 36196 36197
Attributes:
long_name: Raw backscatter measurements (real part)
units: dB- channel: 5
- ping_time: 213
- range_sample: 36198
- 10.38 13.88 16.02 17.17 17.66 17.83 17.86 ... nan nan nan nan nan nan
array([[[ 10.383183 , 13.875601 , 16.015736 , ..., nan, nan, nan], [ 10.430219 , 13.910878 , 16.051014 , ..., nan, nan, nan], [ 10.383183 , 13.875601 , 16.015736 , ..., nan, nan, nan], ..., [ 10.465496 , 13.969673 , 16.121567 , ..., nan, nan, nan], [ 10.465496 , 13.969673 , 16.121567 , ..., nan, nan, nan], [ 10.430219 , 13.899119 , 16.051014 , ..., nan, nan, nan]], [[ -11.782502 , -5.7031074 , -0.50563633, ..., -125.868164 , -126.09158 , -126.29149 ], [ -11.711948 , -5.6443124 , -0.47035936, ..., -133.06467 , -130.23074 , -127.82016 ], [ -11.770743 , -5.6795893 , -0.49387732, ..., -133.14697 , -132.48848 , -131.8182 ], ... [ 3.5394542 , 10.41846 , 15.780557 , ..., nan, nan, nan], [ 3.4689002 , 10.383183 , 15.757039 , ..., nan, nan, nan], [ 3.4924183 , 10.383183 , 15.757039 , ..., nan, nan, nan]], [[ 6.067636 , 8.866274 , 10.747711 , ..., nan, nan, nan], [ 6.079395 , 8.842756 , 10.747711 , ..., nan, nan, nan], [ 6.2087436 , 9.019141 , 10.912337 , ..., nan, nan, nan], ..., [ 6.3028154 , 9.077936 , 10.959373 , ..., nan, nan, nan], [ 6.2087436 , 9.0309 , 10.935855 , ..., nan, nan, nan], [ 6.3028154 , 9.077936 , 10.971132 , ..., nan, nan, nan]]], dtype=float32) - channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - ping_time(ping_time)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
- long_name :
- Timestamp of each ping
- standard_name :
- time
- axis :
- T
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:30.276106000', '2023-08-11T16:57:33.288034000', ..., '2023-08-11T17:05:25.278637000', '2023-08-11T17:05:27.281294000', '2023-08-11T17:05:29.279934000'], dtype='datetime64[ns]') - range_sample(range_sample)int640 1 2 3 ... 36194 36195 36196 36197
- long_name :
- Along-range sample number, base 0
array([ 0, 1, 2, ..., 36195, 36196, 36197])
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - ping_timePandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:30.276106', '2023-08-11 16:57:33.288034', '2023-08-11 16:57:36.293041', '2023-08-11 16:57:39.277018', '2023-08-11 16:57:42.277037', '2023-08-11 16:57:44.287659', '2023-08-11 16:57:46.277308', '2023-08-11 16:57:48.287941', '2023-08-11 16:57:50.278605', ... '2023-08-11 17:05:11.287060', '2023-08-11 17:05:13.278721', '2023-08-11 17:05:15.277386', '2023-08-11 17:05:17.278019', '2023-08-11 17:05:19.278667', '2023-08-11 17:05:21.289291', '2023-08-11 17:05:23.292936', '2023-08-11 17:05:25.278637', '2023-08-11 17:05:27.281294', '2023-08-11 17:05:29.279934'], dtype='datetime64[ns]', name='ping_time', length=213, freq=None)) - range_samplePandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, ... 36188, 36189, 36190, 36191, 36192, 36193, 36194, 36195, 36196, 36197], dtype='int64', name='range_sample', length=36198))
- long_name :
- Raw backscatter measurements (real part)
- units :
- dB
# Lazy-loaded: see the entire array size and the chunk size
ed_new["Sonar/Beam_group1"]["backscatter_r"]<xarray.DataArray 'backscatter_r' (channel: 5, ping_time: 213,
range_sample: 36198)> Size: 154MB
dask.array<open_dataset-backscatter_r, shape=(5, 213, 36198), dtype=float32, chunksize=(5, 213, 23474), chunktype=numpy.ndarray>
Coordinates:
* channel (channel) <U25 500B 'WBT 400140-15 ES120-7C_ES' ... 'WBT 40...
* ping_time (ping_time) datetime64[ns] 2kB 2023-08-11T16:57:27.277163 ....
* range_sample (range_sample) int64 290kB 0 1 2 3 ... 36194 36195 36196 36197
Attributes:
long_name: Raw backscatter measurements (real part)
units: dB- channel: 5
- ping_time: 213
- range_sample: 36198
- dask.array<chunksize=(5, 213, 23474), meta=np.ndarray>
Array Chunk Bytes 147.06 MiB 95.37 MiB Shape (5, 213, 36198) (5, 213, 23474) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - ping_time(ping_time)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
- axis :
- T
- long_name :
- Timestamp of each ping
- standard_name :
- time
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:30.276106000', '2023-08-11T16:57:33.288034000', ..., '2023-08-11T17:05:25.278637000', '2023-08-11T17:05:27.281294000', '2023-08-11T17:05:29.279934000'], dtype='datetime64[ns]') - range_sample(range_sample)int640 1 2 3 ... 36194 36195 36196 36197
- long_name :
- Along-range sample number, base 0
array([ 0, 1, 2, ..., 36195, 36196, 36197])
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - ping_timePandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:30.276106', '2023-08-11 16:57:33.288034', '2023-08-11 16:57:36.293041', '2023-08-11 16:57:39.277018', '2023-08-11 16:57:42.277037', '2023-08-11 16:57:44.287659', '2023-08-11 16:57:46.277308', '2023-08-11 16:57:48.287941', '2023-08-11 16:57:50.278605', ... '2023-08-11 17:05:11.287060', '2023-08-11 17:05:13.278721', '2023-08-11 17:05:15.277386', '2023-08-11 17:05:17.278019', '2023-08-11 17:05:19.278667', '2023-08-11 17:05:21.289291', '2023-08-11 17:05:23.292936', '2023-08-11 17:05:25.278637', '2023-08-11 17:05:27.281294', '2023-08-11 17:05:29.279934'], dtype='datetime64[ns]', name='ping_time', length=213, freq=None)) - range_samplePandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, ... 36188, 36189, 36190, 36191, 36192, 36193, 36194, 36195, 36196, 36197], dtype='int64', name='range_sample', length=36198))
- long_name :
- Raw backscatter measurements (real part)
- units :
- dB
We just need 1 EchoData for processing below, so we can delete ed_new.
Accessing EchoData groups directly via xarray
Note that the zarr or netCDF files on disk are actually standard files, and can be access using regular xarray functions. For example, we an access the GPS data stored in the Platform group directly using xarray.open_dataset:
<xarray.Dataset> Size: 127kB
Dimensions: (channel: 5, time2: 1837, time1: 1451, time3: 1)
Coordinates:
* channel (channel) <U25 500B 'WBT 400140-15 ES120-7C_...
* time1 (time1) datetime64[ns] 12kB 2023-08-11T16:57...
* time2 (time2) datetime64[ns] 15kB 2023-08-11T16:57...
* time3 (time3) datetime64[ns] 8B 2023-08-11T16:57:2...
Data variables: (12/26)
MRU_offset_x float64 8B ...
MRU_offset_y float64 8B ...
MRU_offset_z float64 8B ...
MRU_rotation_x float64 8B ...
MRU_rotation_y float64 8B ...
MRU_rotation_z float64 8B ...
... ...
transducer_offset_x (channel) float64 40B ...
transducer_offset_y (channel) float64 40B ...
transducer_offset_z (channel) float64 40B ...
vertical_offset (time2) float64 15kB ...
water_level float64 8B ...
water_level_draft_is_manual int64 8B ...
Attributes:
platform_code_ICES:
platform_name:
platform_type: - channel: 5
- time2: 1837
- time1: 1451
- time3: 1
- channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - time1(time1)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
- axis :
- T
- comment :
- Time coordinate corresponding to NMEA position data.
- long_name :
- Timestamps for NMEA datagrams
- standard_name :
- time
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', '2023-08-11T16:57:27.277163000', ..., '2023-08-11T17:05:30.095901000', '2023-08-11T17:05:29.858971000', '2023-08-11T17:05:30.086050000'], dtype='datetime64[ns]') - time2(time2)datetime64[ns]2023-08-11T16:57:28.465496 ... 2...
- axis :
- T
- comment :
- Time coordinate corresponding to platform motion and orientation data.
- long_name :
- Timestamps for platform motion and orientation data
- standard_name :
- time
array(['2023-08-11T16:57:28.465496000', '2023-08-11T16:57:28.739187000', '2023-08-11T16:57:28.953922000', ..., '2023-08-11T17:05:30.767675000', '2023-08-11T17:05:31.043379000', '2023-08-11T17:05:31.258127000'], dtype='datetime64[ns]') - time3(time3)datetime64[ns]2023-08-11T16:57:27.277163
- axis :
- T
- comment :
- Time coordinate corresponding to platform motion and orientation data from the Kongsberg Maritime Binary Datagram.
- long_name :
- Timestamps for platform motion and orientation data from the Kongsberg Maritime Binary Datagram
- standard_name :
- time
array(['2023-08-11T16:57:27.277163000'], dtype='datetime64[ns]')
- MRU_offset_x()float64...
- long_name :
- Distance along the x-axis from the platform coordinate system origin to the motion reference unit sensor origin
- units :
- m
[1 values with dtype=float64]
- MRU_offset_y()float64...
- long_name :
- Distance along the y-axis from the platform coordinate system origin to the motion reference unit sensor origin
- units :
- m
[1 values with dtype=float64]
- MRU_offset_z()float64...
- long_name :
- Distance along the z-axis from the platform coordinate system origin to the motion reference unit sensor origin
- units :
- m
[1 values with dtype=float64]
- MRU_rotation_x()float64...
- long_name :
- Extrinsic rotation about the x-axis from the platform to MRU coordinate systems
- units :
- arc_degree
- valid_range :
- (–180.0, 180.0)
[1 values with dtype=float64]
- MRU_rotation_y()float64...
- long_name :
- Extrinsic rotation about the y-axis from the platform to MRU coordinate systems
- units :
- arc_degree
- valid_range :
- (–180.0, 180.0)
[1 values with dtype=float64]
- MRU_rotation_z()float64...
- long_name :
- Extrinsic rotation about the z-axis from the platform to MRU coordinate systems
- units :
- arc_degree
- valid_range :
- (–180.0, 180.0)
[1 values with dtype=float64]
- drop_keel_offset()float64...
[1 values with dtype=float64]
- drop_keel_offset_is_manual()int64...
[1 values with dtype=int64]
- frequency_nominal(channel)float64...
- long_name :
- Transducer frequency
- standard_name :
- sound_frequency
- units :
- Hz
- valid_min :
- 0.0
[5 values with dtype=float64]
- heading(time2)float64...
- long_name :
- Platform heading (true)
- standard_name :
- platform_orientation
- units :
- degrees_north
- valid_max :
- 360.0
- valid_min :
- 0.0
[1837 values with dtype=float64]
- latitude(time1)float64...
- long_name :
- Platform latitude
- standard_name :
- latitude
- units :
- degrees_north
- valid_range :
- (-90.0, 90.0)
[1451 values with dtype=float64]
- latitude_mru1(time3)float64...
- comment :
- Derived from the Simrad MRU1 Datagrams which are a wrapper of the Kongsberg Maritime Binary Datagrams.
- long_name :
- Platform latitude
- standard_name :
- latitude
- units :
- degrees_north
- valid_range :
- (-90.0, 90.0)
[1 values with dtype=float64]
- longitude(time1)float64...
- long_name :
- Platform longitude
- standard_name :
- longitude
- units :
- degrees_east
- valid_range :
- (-180.0, 180.0)
[1451 values with dtype=float64]
- longitude_mru1(time3)float64...
- comment :
- Derived from the Simrad MRU1 Datagrams which are a wrapper of the Kongsberg Maritime Binary Datagrams.
- long_name :
- Platform longitude
- standard_name :
- longitude
- units :
- degrees_east
- valid_range :
- (-180.0, 180.0)
[1 values with dtype=float64]
- pitch(time2)float64...
- long_name :
- Platform pitch
- standard_name :
- platform_pitch_angle
- units :
- arc_degree
- valid_range :
- (-90.0, 90.0)
[1837 values with dtype=float64]
- position_offset_x()float64...
- long_name :
- Distance along the x-axis from the platform coordinate system origin to the latitude/longitude sensor origin
- units :
- m
[1 values with dtype=float64]
- position_offset_y()float64...
- long_name :
- Distance along the y-axis from the platform coordinate system origin to the latitude/longitude sensor origin
- units :
- m
[1 values with dtype=float64]
- position_offset_z()float64...
- long_name :
- Distance along the z-axis from the platform coordinate system origin to the latitude/longitude sensor origin
- units :
- m
[1 values with dtype=float64]
- roll(time2)float64...
- long_name :
- Platform roll
- standard_name :
- platform_roll_angle
- units :
- arc_degree
- valid_range :
- (-90.0, 90.0)
[1837 values with dtype=float64]
- sentence_type(time1)<U3...
- long_name :
- NMEA sentence type
[1451 values with dtype=<U3]
- transducer_offset_x(channel)float64...
- long_name :
- x-axis distance from the platform coordinate system origin to the sonar transducer
- units :
- m
[5 values with dtype=float64]
- transducer_offset_y(channel)float64...
- long_name :
- y-axis distance from the platform coordinate system origin to the sonar transducer
- units :
- m
[5 values with dtype=float64]
- transducer_offset_z(channel)float64...
- long_name :
- z-axis distance from the platform coordinate system origin to the sonar transducer
- units :
- m
[5 values with dtype=float64]
- vertical_offset(time2)float64...
- long_name :
- Platform vertical offset from nominal water level
- units :
- m
[1837 values with dtype=float64]
- water_level()float64...
- long_name :
- Distance from the platform coordinate system origin to the nominal water level along the z-axis
- units :
- m
[1 values with dtype=float64]
- water_level_draft_is_manual()int64...
[1 values with dtype=int64]
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - time1PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:27.277163', '2023-08-11 16:57:28.084934', '2023-08-11 16:57:28.313240', '2023-08-11 16:57:28.917156', '2023-08-11 16:57:29.089186', '2023-08-11 16:57:29.349422', '2023-08-11 16:57:30.080532', '2023-08-11 16:57:29.869759', ... '2023-08-11 17:05:27.311052', '2023-08-11 17:05:28.082108', '2023-08-11 17:05:27.919565', '2023-08-11 17:05:28.302895', '2023-08-11 17:05:28.919241', '2023-08-11 17:05:29.088322', '2023-08-11 17:05:29.324667', '2023-08-11 17:05:30.095901', '2023-08-11 17:05:29.858971', '2023-08-11 17:05:30.086050'], dtype='datetime64[ns]', name='time1', length=1451, freq=None)) - time2PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:28.465496', '2023-08-11 16:57:28.739187', '2023-08-11 16:57:28.953922', '2023-08-11 16:57:29.260298', '2023-08-11 16:57:29.568590', '2023-08-11 16:57:29.844364', '2023-08-11 16:57:30.059101', '2023-08-11 16:57:30.366383', '2023-08-11 16:57:30.642157', '2023-08-11 16:57:30.856892', ... '2023-08-11 17:05:28.957245', '2023-08-11 17:05:29.173046', '2023-08-11 17:05:29.448530', '2023-08-11 17:05:29.665287', '2023-08-11 17:05:29.970560', '2023-08-11 17:05:30.246332', '2023-08-11 17:05:30.461327', '2023-08-11 17:05:30.767675', '2023-08-11 17:05:31.043379', '2023-08-11 17:05:31.258127'], dtype='datetime64[ns]', name='time2', length=1837, freq=None)) - time3PandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163'], dtype='datetime64[ns]', name='time3', freq=None))
- platform_code_ICES :
- platform_name :
- platform_type :
Compute Sv
We can then proceed to compute Sv from the raw data. Here, we will use the calibration and environmental parameters already stored in the original instrument-generated .raw file to do it. Echopype also supports updating these parameters as part of the function call, or use an Echoview .ECS file (though note that this functionality is in beta and may not cover all the equivalent vocabulary recognized by Echoview).
compute_Sv requires two arguments: waveform_mode and encode_mode, because EK80 files can contain data from a variety of sampling configurations, including: - narrowband data stored in power-angle format (waveform_mode="CW", encode_mode="power") - narrowband data stored in complex format (waveform_mode="CW", encode_mode="complex") - broadband data stored in complex format (waveform_mode="FM", encode_mode="complex")
For AZFP echosounder, the arguments will always be waveform_mode="CW", encode_mode="power" since the instrument does not yet support broadband transmissions.
If these arguments are not provided, compute_Sv will error out for EK80 data.
Content of the Sv dataset
<xarray.Dataset> Size: 617MB
Dimensions: (channel: 5, ping_time: 213,
range_sample: 36198, filenames: 1)
Coordinates:
* channel (channel) <U25 500B 'WBT 400140-15 ES120-7...
* ping_time (ping_time) datetime64[ns] 2kB 2023-08-11T...
* range_sample (range_sample) int64 290kB 0 1 ... 36197
* filenames (filenames) int64 8B 0
Data variables: (12/24)
Sv (channel, ping_time, range_sample) float64 308MB ...
echo_range (channel, ping_time, range_sample) float64 308MB ...
frequency_nominal (channel) float64 40B 1.2e+05 ... 2e+05
sound_speed float64 8B 1.48e+03
sound_absorption (channel) float64 40B 0.03281 ... 0.0463
temperature float64 8B 6.9
... ...
beamwidth_athwartship (channel) float64 40B 6.74 10.6 ... 6.81 6.07
impedance_transducer (channel) float64 40B 75.0 75.0 ... 75.0 75.0
impedance_transceiver (channel) int64 40B 5400 5400 5400 5400 5400
receiver_sampling_frequency (channel) float64 40B 1.5e+06 ... 1.5e+06
source_filenames (filenames) <U82 328B 's3://noaa-wcsd-pds/...
water_level float64 8B 0.0
Attributes:
processing_software_name: echopype
processing_software_version: 0.10.1
processing_time: 2025-04-25T18:19:21Z
processing_function: calibrate.compute_Sv- channel: 5
- ping_time: 213
- range_sample: 36198
- filenames: 1
- channel(channel)<U25'WBT 400140-15 ES120-7C_ES' ... ...
- long_name :
- Vendor channel ID
array(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='<U25') - ping_time(ping_time)datetime64[ns]2023-08-11T16:57:27.277163 ... 2...
array(['2023-08-11T16:57:27.277163000', '2023-08-11T16:57:30.276106000', '2023-08-11T16:57:33.288034000', ..., '2023-08-11T17:05:25.278637000', '2023-08-11T17:05:27.281294000', '2023-08-11T17:05:29.279934000'], dtype='datetime64[ns]') - range_sample(range_sample)int640 1 2 3 ... 36194 36195 36196 36197
- long_name :
- Along-range sample number, base 0
array([ 0, 1, 2, ..., 36195, 36196, 36197])
- filenames(filenames)int640
- long_name :
- Index for data and metadata source filenames
array([0])
- Sv(channel, ping_time, range_sample)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Volume backscattering strength (Sv re 1 m-1)
- units :
- dB
- waveform_mode :
- CW
- encode_mode :
- power
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., -78.24315362, -78.46621498, -78.66576255], [ nan, nan, nan, ..., -85.43965631, -82.60537514, -80.19443382], [ nan, nan, nan, ..., -85.52196222, -84.86311134, -84.19248069], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - echo_range(channel, ping_time, range_sample)float640.0 0.0296 0.0592 ... nan nan nan
- long_name :
- Range distance
- units :
- m
array([[[0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan], [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan], [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan], ..., [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan], [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan], [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan]], [[0.00000000e+00, 2.07193286e-02, 4.14386571e-02, ..., 7.49936097e+02, 7.49956817e+02, 7.49977536e+02], [0.00000000e+00, 2.07193286e-02, 4.14386571e-02, ..., 7.49936097e+02, 7.49956817e+02, 7.49977536e+02], [0.00000000e+00, 2.07193286e-02, 4.14386571e-02, ..., 7.49936097e+02, 7.49956817e+02, 7.49977536e+02], ... [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan], [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan], [0.00000000e+00, 2.95990408e-02, 5.91980816e-02, ..., nan, nan, nan]], [[0.00000000e+00, 2.36792326e-02, 4.73584653e-02, ..., nan, nan, nan], [0.00000000e+00, 2.36792326e-02, 4.73584653e-02, ..., nan, nan, nan], [0.00000000e+00, 2.36792326e-02, 4.73584653e-02, ..., nan, nan, nan], ..., [0.00000000e+00, 2.36792326e-02, 4.73584653e-02, ..., nan, nan, nan], [0.00000000e+00, 2.36792326e-02, 4.73584653e-02, ..., nan, nan, nan], [0.00000000e+00, 2.36792326e-02, 4.73584653e-02, ..., nan, nan, nan]]]) - frequency_nominal(channel)float641.2e+05 1.8e+04 7e+04 3.8e+04 2e+05
- units :
- Hz
- long_name :
- Transducer frequency
- valid_min :
- 0.0
- standard_name :
- sound_frequency
array([120000., 18000., 70000., 38000., 200000.])
- sound_speed()float641.48e+03
array(1479.95204001)
- sound_absorption(channel)float640.03281 0.002841 ... 0.0463
array([0.03280678, 0.00284127, 0.02122259, 0.00992475, 0.0463025 ])
- temperature()float646.9
array(6.9)
- salinity()float6433.7
array(33.7)
- pressure()float64200.0
array(200.)
- pH()float648.0
array(8.)
- formula_absorption()<U2'FG'
array('FG', dtype='<U2') - sa_correction(ping_time, channel)float64-0.12 -0.07 -0.2 ... -0.07 -0.09
array([[-0.12, -0.07, -0.2 , -0.07, -0.09], [-0.12, -0.07, -0.2 , -0.07, -0.09], [-0.12, -0.07, -0.2 , -0.07, -0.09], ..., [-0.12, -0.07, -0.2 , -0.07, -0.09], [-0.12, -0.07, -0.2 , -0.07, -0.09], [-0.12, -0.07, -0.2 , -0.07, -0.09]]) - gain_correction(ping_time, channel)float6426.76 23.08 28.1 ... 26.46 27.55
array([[26.76, 23.08, 28.1 , 26.46, 27.55], [26.76, 23.08, 28.1 , 26.46, 27.55], [26.76, 23.08, 28.1 , 26.46, 27.55], ..., [26.76, 23.08, 28.1 , 26.46, 27.55], [26.76, 23.08, 28.1 , 26.46, 27.55], [26.76, 23.08, 28.1 , 26.46, 27.55]]) - equivalent_beam_angle(channel)float64-20.7 -17.0 -20.7 -20.7 -20.7
- long_name :
- Equivalent beam angle
- units :
- sr
- valid_range :
- (0.0, 12.566370614359172)
array([-20.7, -17. , -20.7, -20.7, -20.7])
- angle_offset_alongship(channel)float64-0.04 -0.11 -0.02 -0.2 -0.07
- long_name :
- electrical alongship angle offset of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The alongship angle corresponds to the minor angle in SONAR-netCDF4 vers 2.
array([-0.04, -0.11, -0.02, -0.2 , -0.07])
- angle_offset_athwartship(channel)float640.17 -0.05 0.0 -0.17 0.25
- long_name :
- electrical athwartship angle offset of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The athwartship angle corresponds to the major angle in SONAR-netCDF4 vers 2.
array([ 0.17, -0.05, 0. , -0.17, 0.25])
- angle_sensitivity_alongship(channel)float6423.0 15.5 23.0 23.0 23.0
- long_name :
- alongship angle sensitivity of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The alongship angle corresponds to the minor angle in SONAR-netCDF4 vers 2.
array([23. , 15.5, 23. , 23. , 23. ])
- angle_sensitivity_athwartship(channel)float6423.0 15.5 23.0 23.0 23.0
- long_name :
- athwartship angle sensitivity of the transducer
- comment :
- Introduced in echopype for Simrad echosounders. The athwartship angle corresponds to the major angle in SONAR-netCDF4 vers 2.
array([23. , 15.5, 23. , 23. , 23. ])
- beamwidth_alongship(channel)float646.63 10.92 6.55 6.71 6.79
- long_name :
- Half power two-way beam width along alongship axis of beam
- units :
- arc_degree
- valid_range :
- (0.0, 360.0)
- comment :
- Introduced in echopype for Simrad echosounders to avoid potential confusion with convention definitions. The alongship angle corresponds to the minor angle in SONAR-netCDF4 vers 2. The convention defines one-way transmit or receive beamwidth (beamwidth_receive_minor and beamwidth_transmit_minor), but Simrad echosounders record two-way beamwidth in the data.
array([ 6.63, 10.92, 6.55, 6.71, 6.79])
- beamwidth_athwartship(channel)float646.74 10.6 6.61 6.81 6.07
- long_name :
- Half power two-way beam width along athwartship axis of beam
- units :
- arc_degree
- valid_range :
- (0.0, 360.0)
- comment :
- Introduced in echopype for Simrad echosounders to avoid potential confusion with convention definitions. The athwartship angle corresponds to the major angle in SONAR-netCDF4 vers 2. The convention defines one-way transmit or receive beamwidth (beamwidth_receive_major and beamwidth_transmit_major), but Simrad echosounders record two-way beamwidth in the data.
array([ 6.74, 10.6 , 6.61, 6.81, 6.07])
- impedance_transducer(channel)float6475.0 75.0 75.0 75.0 75.0
array([75., 75., 75., 75., 75.])
- impedance_transceiver(channel)int645400 5400 5400 5400 5400
- units :
- ohm
- long_name :
- Transceiver impedance
array([5400, 5400, 5400, 5400, 5400])
- receiver_sampling_frequency(channel)float641.5e+06 1.5e+06 ... 1.5e+06 1.5e+06
- units :
- Hz
- long_name :
- Receiver sampling frequency
array([1500000., 1500000., 1500000., 1500000., 1500000.])
- source_filenames(filenames)<U82's3://noaa-wcsd-pds/data/raw/Bel...
- long_name :
- Source filenames
array(['s3://noaa-wcsd-pds/data/raw/Bell_M._Shimada/SH2306/EK80/Hake-D20230811-T165727.raw'], dtype='<U82') - water_level()float640.0
- long_name :
- Distance from the platform coordinate system origin to the nominal water level along the z-axis
- units :
- m
array(0.)
- channelPandasIndex
PandasIndex(Index(['WBT 400140-15 ES120-7C_ES', 'WBT 400141-15 ES18_ES', 'WBT 400142-15 ES70-7C_ES', 'WBT 400143-15 ES38B_ES', 'WBT 400145-15 ES200-7C_ES'], dtype='object', name='channel')) - ping_timePandasIndex
PandasIndex(DatetimeIndex(['2023-08-11 16:57:27.277163', '2023-08-11 16:57:30.276106', '2023-08-11 16:57:33.288034', '2023-08-11 16:57:36.293041', '2023-08-11 16:57:39.277018', '2023-08-11 16:57:42.277037', '2023-08-11 16:57:44.287659', '2023-08-11 16:57:46.277308', '2023-08-11 16:57:48.287941', '2023-08-11 16:57:50.278605', ... '2023-08-11 17:05:11.287060', '2023-08-11 17:05:13.278721', '2023-08-11 17:05:15.277386', '2023-08-11 17:05:17.278019', '2023-08-11 17:05:19.278667', '2023-08-11 17:05:21.289291', '2023-08-11 17:05:23.292936', '2023-08-11 17:05:25.278637', '2023-08-11 17:05:27.281294', '2023-08-11 17:05:29.279934'], dtype='datetime64[ns]', name='ping_time', length=213, freq=None)) - range_samplePandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, ... 36188, 36189, 36190, 36191, 36192, 36193, 36194, 36195, 36196, 36197], dtype='int64', name='range_sample', length=36198)) - filenamesPandasIndex
PandasIndex(Index([0], dtype='int64', name='filenames'))
- processing_software_name :
- echopype
- processing_software_version :
- 0.10.1
- processing_time :
- 2025-04-25T18:19:21Z
- processing_function :
- calibrate.compute_Sv
ds_Sv is a standard xarray Dataset with multiple data variable, each an xarray DataArray.
In addition to the Sv variable, there are many other variables in this dataset, including: - all calibration and environmental parameters used to compute the Sv values, to ensure traceability and reproducibility - the echo_range variable that contains the range of each echo sample from the transducer
echo_range has the same dimension as the Sv variable, because the sampling rate of the echo time series (step size along range) and sound speed may vary across pings depending on the echosounder configuration and the environment.
It is also often useful to have the Sv dataset aligned with the frequency_nominal instead of channel:
Let’s take a quick look at what the Sv data looks like:
ds_Sv["Sv"].plot(
x="ping_time",
row="channel", col_wrap=3,
vmin=-80, vmax=-30,
cmap="RdYlBu_r", yincrease=False
)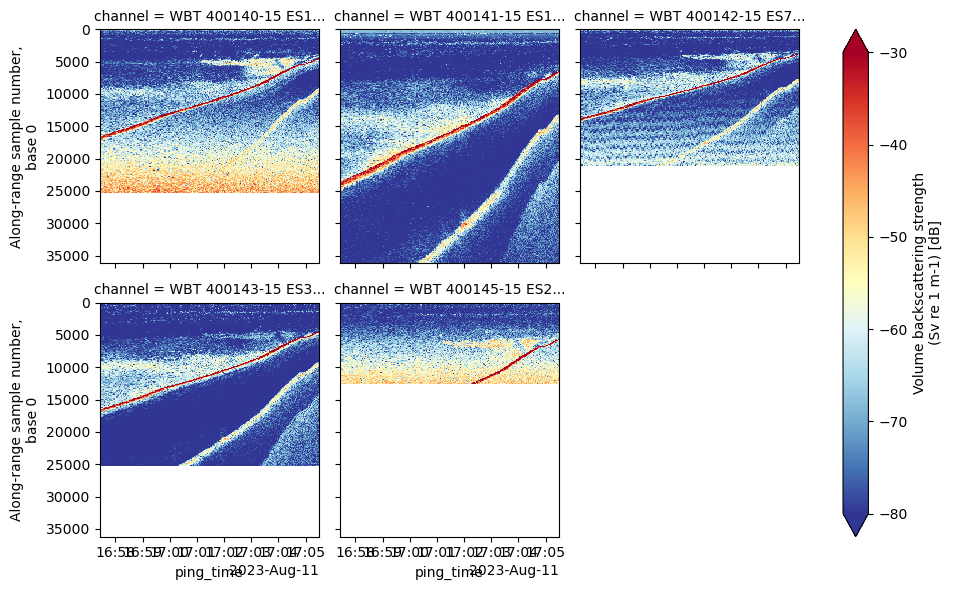
We can see that there are lots of NaN (empty) entries in this dataset, and the number of along-range samples are different for different channels. This is because the echosounder was configured to collect data up to different range, and the sample interval (step size) can also be different for different channels.
In Echopype, we chose to pad NaN for shorter pings and channels, so that the data are gridded at the raw data and all subsequent stages. This makes it much easier to slice different parts of data out, and helps with computing efficiency.
This choice is one of the key differences between Echopype-adapted data format and SONAR-netCDF4 definitions. See the Echopype documentation for more information.
Correct reversed ping time
For some reason, the EK echosounders sometimes generate ping times that are reversed in time, by a tiny amount (<1 msec scale). Reversed coordinate values are problematic for xarray and many operations, and we can correct them by using functions in the qc subpackage:
Add additional information to the Sv dataset
The Sv dataset generated by compute_Sv only contains backscatter data, but we often need additional information, such as the latitude/longitutude location of the platform, or the split-beam angle inferred from the echo time series. We can add these into the dataset by calling functions from the consolidate subpackage. We can also compute the depth of each echo sample if the platform depth is known.
For data saved in complex format, the split-beam angles are calculated from the samplesy. For data saved in power-angle format, the split-beam angle is directly extracted from the EchoData object since it’s already part of the raw file.
Echopype does not yet support heave compensation.
Adding depth and split-beam angle would run through fine:
However, for this particular file, we know that if we try to add the GPS location directly would error out, because there’s are duplicated values in time1.
Therefore, below we first do some cleaning by subsetting only the “GGA” NMEA sentences that have high resolution GPS data and removing the first element that duplicates with the second element in time1.
We decided to not handle problematic GPS data automatically, because it is hard to infer why such data came about in the first place and how to correct it. We feel it is safer for users to make their own decision and clean up their GPS data before proceeding.
After these operations, we now have an Sv dataset that contains pretty useful data that we can use for downstream analysis.
Compute MVBS
We can bring them onto the same grid by computing the mean volume backscattering strength (MVBS):
Now, all channels are bin-averaged on the same grid:
ds_MVBS["Sv"].plot(
x="ping_time",
row="frequency_nominal", col_wrap=3,
cmap='RdYlBu_r', yincrease=False,
vmin=-85, vmax=-45
)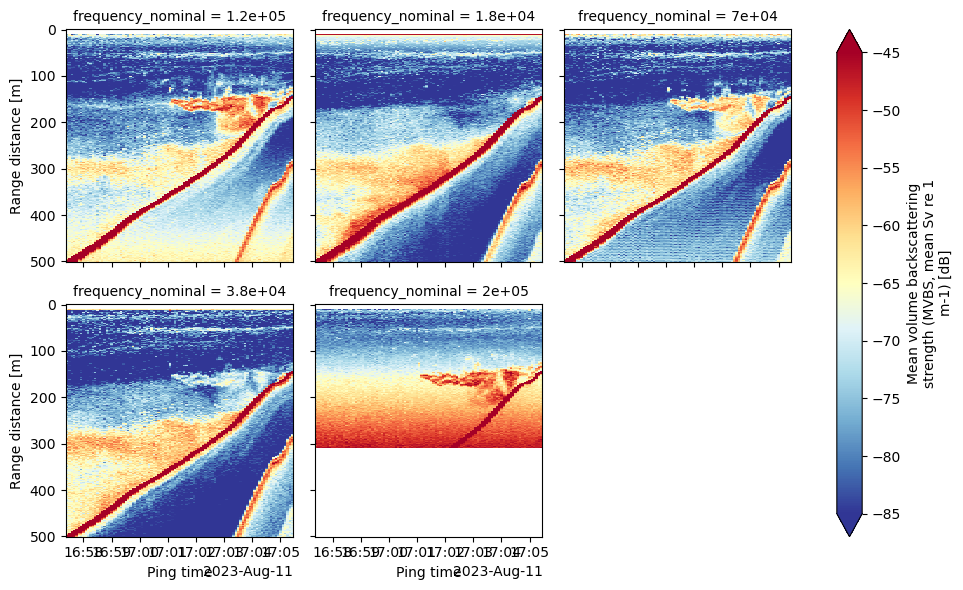
And we can use hvPlot to quickly plot an interactive figure:



