📘 Learning Objectives
Show how to work with the earthaccess package for PACE data
Create a NASA EDL session for authentication
Load single files with xarray.open_dataset
Load multiple files with xarray.open_mfdataset
Overview
The PACE Level-3 (gridded) OCI (ocean color instrument) data is available on an NASA EarthData. Search using the instrument filter “OCI” and processing level filter “Gridded Observations” https://search.earthdata.nasa.gov/search?fi=OCI&fl=3%2B-%2BGridded%2BObservations and you will see 45+ data collections. In this tutorial, we will look at the Spectral phytoplankton absorption coefficients (APH) product.
APH is in the IOP collection: PACE OCI Level-3 Global Mapped Inherent Optical Properties (IOP) Data, version 3.0 . The concept id for this dataset is “C3385050632-OB_CLOUD” and the short name is “PACE_OCI_L3M_IOP”.
Prerequisites
You need to have an EarthData Login username and password. Go here to get one https://urs.earthdata.nasa.gov/
I assume you have a .netrc file at ~ (home). ~/.netrc should look just like this with your username and password. Create that file if needed. You don’t need to create it if you don’t have this file. The earthaccess.login(persist=True) line will ask for your username and password and create the .netrc file for you.
machine urs.earthdata.nasa.gov
login yourusername
password yourpassword
For those not working in the JupyterHub
Uncomment this line and run the cell:
# pip install earthaccess
Create a NASA EDL authenticated session
Authenticate with earthaccess.login().Authenticate with earthaccess.login(). You will need your EarthData Login username and password for this step. Get one here https://urs.earthdata.nasa.gov/ .
import earthaccess= earthaccess.login()# are we authenticated? if not auth.authenticated:# ask for credentials and persist them in a .netrc file = "interactive" , persist= True )
Monthly data
I poked around on the files on search.earthdata so I know what the files look like. March to December 2024.
import earthaccess= earthaccess.search_data(= "PACE_OCI_L3M_IOP" ,= ("2024-03-01" , "2024-12-31" ),= "*.MO.*aph.0p1deg.*" len (results_mo)
# Create a fileset = earthaccess.open (results_mo);
# let's load just one month import xarray as xr= xr.open_dataset(fileset[0 ])
<xarray.Dataset> Size: 493MB
Dimensions: (wavelength: 19, lat: 1800, lon: 3600, rgb: 3,
eightbitcolor: 256)
Coordinates:
* wavelength (wavelength) float64 152B 351.0 361.0 385.0 ... 678.0 711.0
* lat (lat) float32 7kB 89.95 89.85 89.75 ... -89.75 -89.85 -89.95
* lon (lon) float32 14kB -179.9 -179.9 -179.8 ... 179.8 179.9 180.0
Dimensions without coordinates: rgb, eightbitcolor
Data variables:
aph (lat, lon, wavelength) float32 492MB ...
palette (rgb, eightbitcolor) uint8 768B ...
Attributes: (12/64)
product_name: PACE_OCI.20240301_20240331.L3m.MO.IOP....
instrument: OCI
title: OCI Level-3 Standard Mapped Image
project: Ocean Biology Processing Group (NASA/G...
platform: PACE
source: satellite observations from OCI-PACE
... ...
identifier_product_doi: 10.5067/PACE/OCI/L3M/IOP/3.0
keywords: Earth Science > Oceans > Ocean Optics ...
keywords_vocabulary: NASA Global Change Master Directory (G...
data_bins: 3016341
data_minimum: -0.7766001
data_maximum: 5.0000014 Dimensions: wavelength : 19lat : 1800lon : 3600rgb : 3eightbitcolor : 256
Coordinates: (3)
wavelength
(wavelength)
float64
351.0 361.0 385.0 ... 678.0 711.0
long_name : wavelengths units : nm valid_min : 0 valid_max : 20000 array([351., 361., 385., 413., 425., 442., 460., 475., 490., 510., 532., 555.,
583., 618., 640., 655., 665., 678., 711.]) lat
(lat)
float32
89.95 89.85 89.75 ... -89.85 -89.95
long_name : Latitude units : degrees_north standard_name : latitude valid_min : -90.0 valid_max : 90.0 array([ 89.95 , 89.85 , 89.75 , ..., -89.75 , -89.850006,
-89.950005], dtype=float32) lon
(lon)
float32
-179.9 -179.9 ... 179.9 180.0
long_name : Longitude units : degrees_east standard_name : longitude valid_min : -180.0 valid_max : 180.0 array([-179.95 , -179.85 , -179.75 , ..., 179.75 , 179.85 ,
179.95001], dtype=float32) Data variables: (2)
Indexes: (3)
PandasIndex
PandasIndex(Index([351.0, 361.0, 385.0, 413.0, 425.0, 442.0, 460.0, 475.0, 490.0, 510.0,
532.0, 555.0, 583.0, 618.0, 640.0, 655.0, 665.0, 678.0, 711.0],
dtype='float64', name='wavelength')) PandasIndex
PandasIndex(Index([ 89.94999694824219, 89.8499984741211, 89.75,
89.6500015258789, 89.55000305175781, 89.44999694824219,
89.3499984741211, 89.25, 89.1500015258789,
89.05000305175781,
...
-89.05000305175781, -89.1500015258789, -89.25,
-89.35000610351562, -89.45000457763672, -89.55000305175781,
-89.6500015258789, -89.75, -89.85000610351562,
-89.95000457763672],
dtype='float32', name='lat', length=1800)) PandasIndex
PandasIndex(Index([ -179.9499969482422, -179.85000610351562, -179.75,
-179.64999389648438, -179.5500030517578, -179.4499969482422,
-179.35000610351562, -179.25, -179.14999389648438,
-179.0500030517578,
...
179.0500030517578, 179.15000915527344, 179.25,
179.35000610351562, 179.45001220703125, 179.5500030517578,
179.65000915527344, 179.75, 179.85000610351562,
179.95001220703125],
dtype='float32', name='lon', length=3600)) Attributes: (64)
product_name : PACE_OCI.20240301_20240331.L3m.MO.IOP.V3_0.aph.0p1deg.nc instrument : OCI title : OCI Level-3 Standard Mapped Image project : Ocean Biology Processing Group (NASA/GSFC/OBPG) platform : PACE source : satellite observations from OCI-PACE temporal_range : 27-day processing_version : 3.0 date_created : 2025-03-06T17:36:25.000Z history : l3mapgen par=PACE_OCI.20240301_20240331.L3m.MO.IOP.V3_0.aph.0p1deg.nc.param l2_flag_names : ATMFAIL,LAND,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLWARN,CHLFAIL,NAVWARN,MAXAERITER,HISOLZEN,NAVFAIL,FILTER,HIGLINT time_coverage_start : 2024-03-05T00:08:58.000Z time_coverage_end : 2024-04-01T02:24:44.000Z start_orbit_number : 0 end_orbit_number : 0 map_projection : Equidistant Cylindrical latitude_units : degrees_north longitude_units : degrees_east northernmost_latitude : 90.0 southernmost_latitude : -90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 geospatial_lat_max : 90.0 geospatial_lat_min : -90.0 geospatial_lon_max : 180.0 geospatial_lon_min : -180.0 latitude_step : 0.1 longitude_step : 0.1 sw_point_latitude : -89.95 sw_point_longitude : -179.95 spatialResolution : 11.131949 km geospatial_lon_resolution : 11.131949 km geospatial_lat_resolution : 11.131949 km geospatial_lat_units : degrees_north geospatial_lon_units : degrees_east number_of_lines : 1800 number_of_columns : 3600 measure : Mean suggested_image_scaling_minimum : 0.001 suggested_image_scaling_maximum : 1.0 suggested_image_scaling_type : LOG suggested_image_scaling_applied : No _lastModified : 2025-03-06T17:36:25.000Z Conventions : CF-1.6 ACDD-1.3 institution : NASA Goddard Space Flight Center, Ocean Ecology Laboratory, Ocean Biology Processing Group standard_name_vocabulary : CF Standard Name Table v36 naming_authority : gov.nasa.gsfc.sci.oceandata id : 3.0/L3/PACE_OCI.20240301_20240331.L3b.MO.IOP.V3_0.nc license : https://science.nasa.gov/earth-science/earth-science-data/data-information-policy/ creator_name : NASA/GSFC/OBPG publisher_name : NASA/GSFC/OBPG creator_email : data@oceancolor.gsfc.nasa.gov publisher_email : data@oceancolor.gsfc.nasa.gov creator_url : https://oceandata.sci.gsfc.nasa.gov publisher_url : https://oceandata.sci.gsfc.nasa.gov processing_level : L3 Mapped cdm_data_type : grid identifier_product_doi_authority : http://dx.doi.org identifier_product_doi : 10.5067/PACE/OCI/L3M/IOP/3.0 keywords : Earth Science > Oceans > Ocean Optics > Reflectance keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords data_bins : 3016341 data_minimum : -0.7766001 data_maximum : 5.0000014
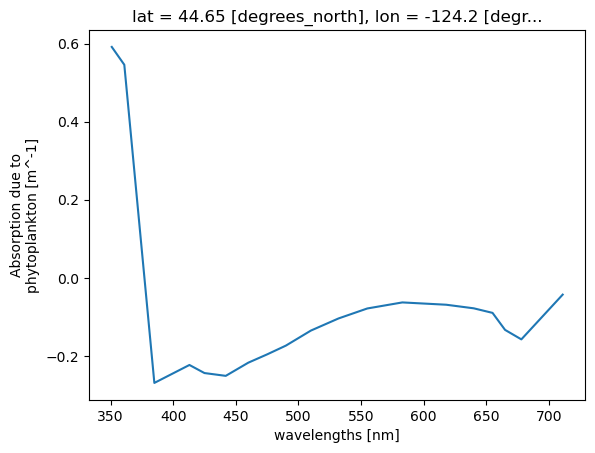
= ds['aph' ].sel(lat= 44.6517 , lon=- 124.1770 , method= 'nearest' );
Multiple months
= xr.open_mfdataset(= 'nested' , concat_dim= "time"
<xarray.Dataset> Size: 5GB
Dimensions: (time: 10, lat: 1800, lon: 3600, wavelength: 19, rgb: 3,
eightbitcolor: 256)
Coordinates:
* wavelength (wavelength) float64 152B 351.0 361.0 385.0 ... 678.0 711.0
* lat (lat) float32 7kB 89.95 89.85 89.75 ... -89.75 -89.85 -89.95
* lon (lon) float32 14kB -179.9 -179.9 -179.8 ... 179.8 179.9 180.0
Dimensions without coordinates: time, rgb, eightbitcolor
Data variables:
aph (time, lat, lon, wavelength) float32 5GB dask.array<chunksize=(1, 16, 1024, 8), meta=np.ndarray>
palette (time, rgb, eightbitcolor) uint8 8kB dask.array<chunksize=(1, 3, 256), meta=np.ndarray>
Attributes: (12/64)
product_name: PACE_OCI.20240301_20240331.L3m.MO.IOP....
instrument: OCI
title: OCI Level-3 Standard Mapped Image
project: Ocean Biology Processing Group (NASA/G...
platform: PACE
source: satellite observations from OCI-PACE
... ...
identifier_product_doi: 10.5067/PACE/OCI/L3M/IOP/3.0
keywords: Earth Science > Oceans > Ocean Optics ...
keywords_vocabulary: NASA Global Change Master Directory (G...
data_bins: 3016341
data_minimum: -0.7766001
data_maximum: 5.0000014 Dimensions: time : 10lat : 1800lon : 3600wavelength : 19rgb : 3eightbitcolor : 256
Coordinates: (3)
wavelength
(wavelength)
float64
351.0 361.0 385.0 ... 678.0 711.0
long_name : wavelengths units : nm valid_min : 0 valid_max : 20000 array([351., 361., 385., 413., 425., 442., 460., 475., 490., 510., 532., 555.,
583., 618., 640., 655., 665., 678., 711.]) lat
(lat)
float32
89.95 89.85 89.75 ... -89.85 -89.95
long_name : Latitude units : degrees_north standard_name : latitude valid_min : -90.0 valid_max : 90.0 array([ 89.95 , 89.85 , 89.75 , ..., -89.75 , -89.850006,
-89.950005], dtype=float32) lon
(lon)
float32
-179.9 -179.9 ... 179.9 180.0
long_name : Longitude units : degrees_east standard_name : longitude valid_min : -180.0 valid_max : 180.0 array([-179.95 , -179.85 , -179.75 , ..., 179.75 , 179.85 ,
179.95001], dtype=float32) Data variables: (2)
aph
(time, lat, lon, wavelength)
float32
dask.array<chunksize=(1, 16, 1024, 8), meta=np.ndarray>
long_name : Absorption due to phytoplankton units : m^-1 valid_min : -24998 valid_max : 25000 reference : P.J. Werdell, B.A. Franz, S.W. Bailey, G.C. Feldman and 15 co-authors, Generalized ocean color inversion model for retrieving marine inherent optical properties, Applied Optics 52, 2019-2037 (2013). display_scale : log display_min : 0.001 display_max : 1.0
Bytes
4.59 GiB
512.00 kiB
Shape
(10, 1800, 3600, 19)
(1, 16, 1024, 8)
Dask graph
13560 chunks in 31 graph layers
Data type
float32 numpy.ndarray
10 1 19 3600 1800
palette
(time, rgb, eightbitcolor)
uint8
dask.array<chunksize=(1, 3, 256), meta=np.ndarray>
Bytes
7.50 kiB
768 B
Shape
(10, 3, 256)
(1, 3, 256)
Dask graph
10 chunks in 31 graph layers
Data type
uint8 numpy.ndarray
256 3 10
Indexes: (3)
PandasIndex
PandasIndex(Index([351.0, 361.0, 385.0, 413.0, 425.0, 442.0, 460.0, 475.0, 490.0, 510.0,
532.0, 555.0, 583.0, 618.0, 640.0, 655.0, 665.0, 678.0, 711.0],
dtype='float64', name='wavelength')) PandasIndex
PandasIndex(Index([ 89.94999694824219, 89.8499984741211, 89.75,
89.6500015258789, 89.55000305175781, 89.44999694824219,
89.3499984741211, 89.25, 89.1500015258789,
89.05000305175781,
...
-89.05000305175781, -89.1500015258789, -89.25,
-89.35000610351562, -89.45000457763672, -89.55000305175781,
-89.6500015258789, -89.75, -89.85000610351562,
-89.95000457763672],
dtype='float32', name='lat', length=1800)) PandasIndex
PandasIndex(Index([ -179.9499969482422, -179.85000610351562, -179.75,
-179.64999389648438, -179.5500030517578, -179.4499969482422,
-179.35000610351562, -179.25, -179.14999389648438,
-179.0500030517578,
...
179.0500030517578, 179.15000915527344, 179.25,
179.35000610351562, 179.45001220703125, 179.5500030517578,
179.65000915527344, 179.75, 179.85000610351562,
179.95001220703125],
dtype='float32', name='lon', length=3600)) Attributes: (64)
product_name : PACE_OCI.20240301_20240331.L3m.MO.IOP.V3_0.aph.0p1deg.nc instrument : OCI title : OCI Level-3 Standard Mapped Image project : Ocean Biology Processing Group (NASA/GSFC/OBPG) platform : PACE source : satellite observations from OCI-PACE temporal_range : 27-day processing_version : 3.0 date_created : 2025-03-06T17:36:25.000Z history : l3mapgen par=PACE_OCI.20240301_20240331.L3m.MO.IOP.V3_0.aph.0p1deg.nc.param l2_flag_names : ATMFAIL,LAND,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLWARN,CHLFAIL,NAVWARN,MAXAERITER,HISOLZEN,NAVFAIL,FILTER,HIGLINT time_coverage_start : 2024-03-05T00:08:58.000Z time_coverage_end : 2024-04-01T02:24:44.000Z start_orbit_number : 0 end_orbit_number : 0 map_projection : Equidistant Cylindrical latitude_units : degrees_north longitude_units : degrees_east northernmost_latitude : 90.0 southernmost_latitude : -90.0 westernmost_longitude : -180.0 easternmost_longitude : 180.0 geospatial_lat_max : 90.0 geospatial_lat_min : -90.0 geospatial_lon_max : 180.0 geospatial_lon_min : -180.0 latitude_step : 0.1 longitude_step : 0.1 sw_point_latitude : -89.95 sw_point_longitude : -179.95 spatialResolution : 11.131949 km geospatial_lon_resolution : 11.131949 km geospatial_lat_resolution : 11.131949 km geospatial_lat_units : degrees_north geospatial_lon_units : degrees_east number_of_lines : 1800 number_of_columns : 3600 measure : Mean suggested_image_scaling_minimum : 0.001 suggested_image_scaling_maximum : 1.0 suggested_image_scaling_type : LOG suggested_image_scaling_applied : No _lastModified : 2025-03-06T17:36:25.000Z Conventions : CF-1.6 ACDD-1.3 institution : NASA Goddard Space Flight Center, Ocean Ecology Laboratory, Ocean Biology Processing Group standard_name_vocabulary : CF Standard Name Table v36 naming_authority : gov.nasa.gsfc.sci.oceandata id : 3.0/L3/PACE_OCI.20240301_20240331.L3b.MO.IOP.V3_0.nc license : https://science.nasa.gov/earth-science/earth-science-data/data-information-policy/ creator_name : NASA/GSFC/OBPG publisher_name : NASA/GSFC/OBPG creator_email : data@oceancolor.gsfc.nasa.gov publisher_email : data@oceancolor.gsfc.nasa.gov creator_url : https://oceandata.sci.gsfc.nasa.gov publisher_url : https://oceandata.sci.gsfc.nasa.gov processing_level : L3 Mapped cdm_data_type : grid identifier_product_doi_authority : http://dx.doi.org identifier_product_doi : 10.5067/PACE/OCI/L3M/IOP/3.0 keywords : Earth Science > Oceans > Ocean Optics > Reflectance keywords_vocabulary : NASA Global Change Master Directory (GCMD) Science Keywords data_bins : 3016341 data_minimum : -0.7766001 data_maximum : 5.0000014
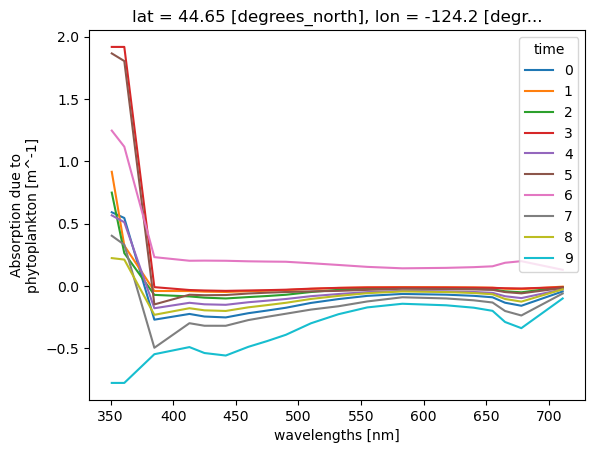
= ds['aph' ].sel(lat= 44.6517 , lon=- 124.1770 , method= 'nearest' )= "wavelength" );
Raster plots of the APH
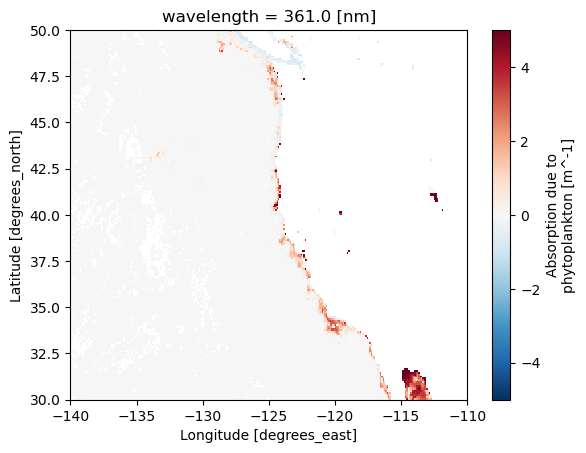
"aph" ].isel(time= 0 ).sel(wavelength = 361 , lat = slice (50 , 30 ), lon= slice (- 140 , - 110 )).plot();
array([351., 361., 385., 413., 425., 442., 460., 475., 490., 510., 532.,
555., 583., 618., 640., 655., 665., 678., 711.])
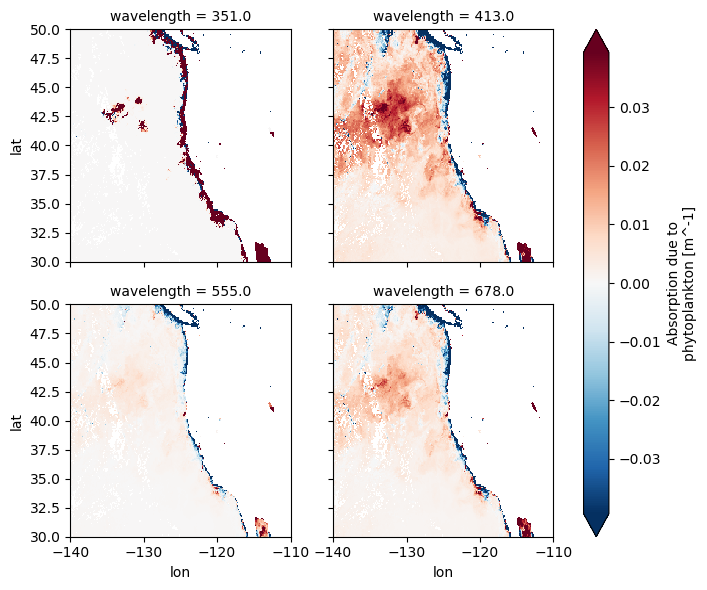
"aph" ].isel(time= 0 ).sel(= slice (50 , 30 ), = slice (- 140 , - 110 ), = [351 , 413 , 555 , 678 ] # choose your wavelengths = "lon" , y= "lat" , col= "wavelength" , col_wrap= 2 , robust= True );